Abstract
Commercially produced ground beef samples (n = 4,136) were collected from seven regions of the United States over a 24-month period (July 2005 to June 2007) and analyzed for the presence of Salmonella enterica by using methods that concurrently provided total prevalence and enumerable levels. The overall prevalence of Salmonella strains was 4.2%. Enumeration showed that 94.2% were present at levels below 2 CFU/g. Regional monthly prevalences of Salmonella strains varied from 1.8% to 6.5% but were not statistically different (P > 0.05). All Salmonella isolates were serotyped and their antibiotic susceptibilities determined and analyzed by pulsed-field gel electrophoresis (PFGE). The most common serotypes identified were Salmonella enterica serotypes Montevideo, Anatum, Muenster, and Mbandaka, with these accounting for one-half of the isolates obtained. The prevalence of multidrug-resistant (MDR) Salmonella was determined to be 0.6%. The most common MDR serotypes were Salmonella enterica serotypes Dublin, Reading, and Typhimurium. MDR strains had resistance to between 2 and 10 antibiotics. There were no regional differences in prevalence of MDR Salmonella. PFGE analysis revealed that indistinguishable XbaI and AvrII restriction digest patterns (RDPs) could be observed in isolates of the same serotype found in different regions and months of sampling. The RDPs of 19 Salmonella strains were compared to RDPs in the PulseNet USA database. Thirteen were indistinguishable from existing patterns, and the number of records for each ranged from 1 to 478. These data show that Salmonella prevalence in commercial ground beef is low and suggest that attempts to identify sources contributing to Salmonella in ground beef by serotype, antibiogram, and PFGE cannot be made without additional documented evidence.
Salmonella enterica bacteria are estimated to cause 1.3 million cases of gastroenteritis each year in the United States (28). Cattle are among the known reservoirs of Salmonella, and ground beef has been implicated as one mode of transmission in food-borne outbreaks (11, 14). The Food Safety Inspection Service (FSIS) reported that Salmonella in ground beef has decreased by 30% (7.5% to 5.8%) with the advent of hazard analysis and critical control point plans at large and small beef processors (18). The current FSIS progress report suggests that the overall prevalence in ground beef is 2.4%. However, the FSIS Salmonella prevalence numbers are based on testing methods (2) that rely solely on enrichment, plating, and biochemical tests without the use of immunomagnetic separation (IMS) to concentrate any Salmonella cells present. The use of IMS has been shown to have a greater level of sensitivity than standard culturing techniques (7). Therefore, the current prevalence of Salmonella in ground beef as presented by FSIS may be an underestimate. In addition, pathogen levels are rarely if ever determined from samples found to be contaminated with Salmonella. The presence of any Salmonella is potentially a health threat, but greater levels of the pathogen carry an increased risk of illness due to the infectious dose of the organism, which varies by strain and host susceptibility (reportedly as low as 15 to 20 cells) (5).
The monitoring and regulation of Salmonella have become more stringent recently (docket no. 4-026N; Federal Register), with the FSIS posting a new approach to verification activities in meat and poultry establishments. These regulations focus on (i) the control of Salmonella entry into the ground beef supply and (ii) the ability to trace the source of the contaminated product. This program relies on information collected on Salmonella serotypes, pulsed-field gel electrophoresis (PFGE) patterns, and antibiograms, coupled with data from the Centers for Disease Control and Prevention (CDC), to enhance source tracking.
The National Antibiotic Resistance Monitoring System (NARMS) has been monitoring retail fresh meats, including ground beef, for Salmonella and determining antibiotic susceptibilities since 1996 (6, 36). The NARMS database is substantial, but the Salmonella information concerning ground beef is overwhelmed by information about Salmonella bacteria that have been isolated from poultry and pork products. Further, the sampling scheme used by the FoodNet laboratories that supply NARMS with isolates examines retail products from a small set of locations. Often, beef, pork, and poultry retail products are handled, packaged, and sold next to one another. The instances for cross-contamination of ground beef in these cases cannot be ruled out. Commercially produced ground beef, as was used in the study presented herein, is regionally produced and distributed to numerous restaurant chains and supermarkets. These samples are unlikely to be handled or exposed to other meat products, and thus, the data collected in this study are more likely to give an accurate description of the Salmonella prevalence in ground beef.
Salmonella strains that are resistant to multiple antibiotics (MDR) have emerged as a food safety concern. An MDR Salmonella enterica serotype Newport strain was responsible for an outbreak linked to ground beef in 2002 causing 47 illnesses, 17 hospitalizations, and one death (14). Typically, only a small percentage of Salmonella strains are resistant to antibiotics, so in order to determine the prevalence of Salmonella in ground beef, a large sample number would be required, and currently, reports in the literature are limited. Of 50 samples of ground beef collected from three stores, 3 (6%) contained Salmonella and 1 (2%) of those 3 was an MDR Salmonella enterica serotype Agona strain (34). In another report of 404 samples from 96 stores, there were 14 (3.5%) Salmonella isolates (37). Five (1.2%) of these isolates were MDR Salmonella enterica serotype Typhimurium DT104 strains that possessed indistinguishable PFGE patterns and came from the same location. These isolates may have been the result of a single undetermined source, but this is uncertain since Salmonella serotype Typhimurium DT104 is very clonal group of Salmonella (21).
Here, we report the results of a 2-year analysis of commercially produced U.S. ground beef from multiple grinding establishments across the country. Prevalence and level of Salmonella were determined. The isolated Salmonella strains were characterized for serotype and antibiotic susceptibility. Additionally, all isolates were subjected to PFGE using two different restriction endonucleases. The PFGE restriction digest patterns (RDPs) were analyzed and compared to those for Salmonella isolates from human disease as reported by the CDC in the PulseNet database.
MATERIALS AND METHODS
Samples.
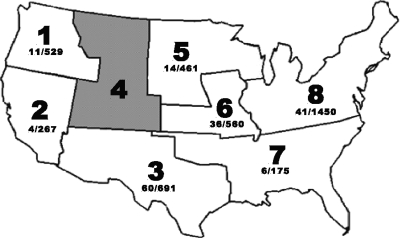
The samples consisted of ground beef collected by 18 commercial ground beef producers that represented a significant cross section of such companies that supply ground beef as patties or case-ready products to numerous restaurant and grocery chains. When the participating producers were organized according to the eight microbiological monitoring regions defined by the Beef Industry Food Safety Council (BIFSCo), one region (region 4, comprising MT, CO, UT, and WY) was not represented (see Fig. 1). The samples ranged from 65 g to 1 kg and had various percentages of lean meat (73%, 80%, 85%, and 90%). The ground beef samples were collected during normal production runs in the form of patties or chubs. Usually, one sample was collected on a given day at a given establishment, and if an establishment collected more than one sample in a day, it was from a separate production run (for example, a product with a different percentage of lean meat). Samples were collected and placed in a Whirl-Pak bag (Nasco, Fort Atchison, WI) and frozen at −20°C (−4°F) for up to 45 days. Approximately once a month, the producers shipped their frozen samples to the U.S. Meat Animal Research Center through an independent third party that removed plant-specific information but provided region and collection date. After receipt, the samples were held at −20°C (−4°F) for up to 15 more days before being processed, resulting in freezing of some samples for up to 60 days. Since the ground beef samples were frozen prior to testing, positive controls were included with each run of samples. The controls were ground beef samples that had been inoculated with Salmonella serotype Typhimurium at a low level of 10 to 20 CFU/65 g to verify isolation procedures and at a higher level of approximately 150 CFU/65 g to verify enumeration procedures and recovery of injured cells. The inoculated controls were frozen at −20°C (−4°F) for 4 to 18 weeks before being thawed and processed with the submitted samples.
FIG. 1.
Map of the BIFSCo microbiological monitoring regions. Ground beef samples were obtained from regions 1, 2, 3, 5, 6, 7, and 8 for this study. Below each region label is the total number of Salmonella-positive samples over the total number of samples received from commercial ground beef producers in that region.
Isolation of Salmonella.
Salmonella strains were isolated from thawed ground beef samples as follows. Each ground beef sample was weighed out (65 g) into a 7.5- by 12-in. filtered Whirl-Pak bag and then thoroughly mixed with 292.5 ml Difco trypticase soy broth (TSB) (Becton Dickinson, Sparks, MD), using a laboratory sample blender (BagMixer 400; Interscience, Weymouth, MA) for 1 min at 420 rpm. A second, 292.5-ml aliquot of TSB was added and thoroughly mixed by hand massaging in lieu of additional stomaching since the laboratory blenders were unable to accommodate volumes over 400 ml. In situations where less than 65 g of ground beef was available, the TSB volume was adjusted to maintain a 1:10 ratio. Five milliliters of the ground beef-TSB dilution was removed from every sample for use in the enumeration assay (described below). The remainder proceeded to incubation as previously described (7) and then was subjected to IMS (1, 16) using 20 μl of Salmonella-specific IMS beads (Dynal, Lake Success, NY). IMS was included in the method of isolation because it provides increased detection and isolation of Salmonella with decreased background organism growth compared to other, traditional methods that use only selective media, such as those described in the FSIS Microbiology Laboratory Guidebook (2). A 1-ml aliquot of each enrichment sample was removed and mixed with IMS beads at room temperature for 15 min with shaking, and then, the bacterium-bead complex was removed using a KingFisher IMS robot (Thermo-Fisher, Waltham, MA), washed twice with phosphate-buffered saline containing 0.5% (vol/vol) Tween 20 (PBS-T; Sigma, St. Louis, MO), and released in 0.1 ml PBS-T. The IMS beads were placed into 3 ml of Rappaport-Vassiliadis soya peptone broth (Oxoid, Basingstoke, United Kingdom) for secondary enrichment and incubated at 42°C for 18 to 20 h. These enrichments were swabbed onto Difco Hektoen enteric medium (Becton Dickinson, Franklin Lakes, NJ) with novobiocin at a concentration of 5 mg liter−1 (HEn) and Difco brilliant green agar with sulfadiazine at 80 mg liter−1 (BGS; Becton Dickinson) and then streaked for isolation. Plates were incubated at 37°C for 18 to 20 h. Black colonies on HEn or pink colonies on BGS were considered putative Salmonella isolates until confirmed by PCR and serotyping. Four suspect colonies (two from each plate when present) were taken forward for confirmation by PCR. When colonies were picked, visual differences in size or morphology were considered, so as to provide the greatest variety of isolates possible. For instance, when large and small black colonies were present on HEn, one of each was picked for subsequent testing. If the situation warranted, additional colonies with various morphologies were picked, and if only one suspect colony was present, it was the only colony processed for that particular sample. The confirmatory PCR method targeted the Salmonella-specific portion of the invA gene (31) and has been validated as a means of accurately identifying isolates of Salmonella enterica (27, 30). The inoculated control ground beef (described above) was included to confirm the performance of the Salmonella isolation and identification procedures.
Enumeration.
Enumeration of Salmonella was performed on all samples by adding 2 ml of phosphate-buffered saline with 1% (vol/vol) Tween 80 (Sigma) to each of the 5-ml aliquots collected from the 1:10 dilutions of ground beef in TSB. This dilution was then vortexed, and debris was allowed to settle for 5 to 10 min and then analyzed by hydrophobic grid membrane filtration (10). Each entire sample, less the sediment, was applied to an ISOGRID membrane (Neogen, Lansing, MI) filtered using a FiltaFlex spread filter apparatus (FiltaFlex, Ltd., Almont, Ontario, Canada), and the membrane was then placed on an XLDtnc agar plate (xylose lysine desoxycholate medium [Oxoid, Remel, Lenexa, KS] with 4.6 ml liter−1 tergitol [Sigma], 15 mg liter−1 novobiocin, and 5 mg liter−1 cefsulodin). The plates were incubated at 37°C for 18 to 20 h and then examined for black colonies, which were presumptively considered Salmonella. The total number of colonies was counted, and up to 10 were selected for evaluation by PCR (described above). When >10 colonies were present, the percentage of those 10 that were confirmed as Salmonella by PCR was multiplied by the total number of colonies counted, and this number was reported in CFU per gram. The inoculated control ground beef (described above) was included to confirm the performance of Salmonella enumeration and the recovery of injured cells.
Antibiotic susceptibility determination.
All Salmonella isolates were screened for resistance to tetracycline (Tet), chloramphenicol, and ampicillin by direct plating then subjected to antibiotic sensitivity panels to determine MICs. Salmonella isolates were stamped in a 96-well block format, using a Boekel microplate replicator, onto tryptic soy agar plates (150 mm) containing either no antibiotics, Tet (32 mg/liter), chloramphenicol (32 mg/liter), or ampicillin (32 mg/liter). These plates were incubated at 37°C for 18 to 20 h. Isolates demonstrating resistance to any of these antibiotics were then streaked for isolation onto tryptic soy agar. Using this scheme, we were able to screen hundreds of isolates for resistance and multidrug resistance in a rapid fashion. The NARMS (6) and others (9, 33, 38) have shown that resistances to these antibiotics are the most frequently observed. The resulting pure cultures were used for antibiotic sensitivity analysis and serological identification. Antibiotic sensitivity was determined by performing MIC tests using the defined NARMS Salmonella antibiotic panels (CMV1AGNF; Trek Diagnostic Systems, Inc., Cleveland, OH) and a Sensititre AutoInoculator and AutoReader (Trek Diagnostic Systems, Inc.). The antibiotics and breakpoints for resistance in this panel were as follows: amoxicillin-clavulanate, ≥32 μg/ml; ampicillin, ≥32 μg/ml; cefoxitin, ≥32 μg/ml; ceftiofur, ≥8 μg/ml; ceftriaxone, ≥64 μg/ml; chloramphenicol, ≥32 μg/ml; gentamicin, ≥16 μg/ml; kanamycin, ≥64 μg/ml; nalidixic acid, ≥32 μg/ml; streptomycin, ≥64 μg/ml; sulfisoxazol, ≥256 μg/ml; and Tet, ≥32 μg/ml. Isolates that had ceftriaxone MICs of 16 to 32 μg/ml or gentamicin MICs of 8 μg/ml were categorized as intermediately resistant to these antibiotics. Results for automated MICs were confirmed using control organisms consisting of a nonresistant Salmonella enterica serotype Anatum strain and a multiply resistant Salmonella serotype Newport strain that had previously been isolated and characterized.
Salmonella serotype determinations.
All confirmed Salmonella isolates were serogrouped using Wellcolex Color Salmonella tests (Remel, Lenexa, KS) according to the manufacturer's recommendations and serotyped (1) using antisera for the identification of somatic and flagellar antigens (Remel). Briefly, slide agglutination was used to confirm or refine the results of the Wellcolex Color test results. This was done by suspending the bacteria (∼5 μl) in saline (0.3 ml) and then mixing 10 μl of this suspension with appropriate O-grouping antisera and control saline on a glass slide, followed by visual observation of agglutination. Following identification of the O group, the first- and second-phase flagella were identified by tube agglutinations. The saline suspension used for O-group identification was used to inoculate a motility agar plate that was incubated for 16 to 40 h at 37°C until swarming bacteria were observed. Swarming bacteria were subcultured in to trypticase soy tryptose broth, grown to mid-log phase, and fixed by addition of formalized saline. The fixed bacteria (30 μl) were tested with H-antigen sera to identify first-phase flagella. After the Salmonella bacteria were phase switched on motility agar that had been treated with specific antisera to the first-phase H antigen, the second-phase flagella were identified in a similar fashion.
PFGE analyses.
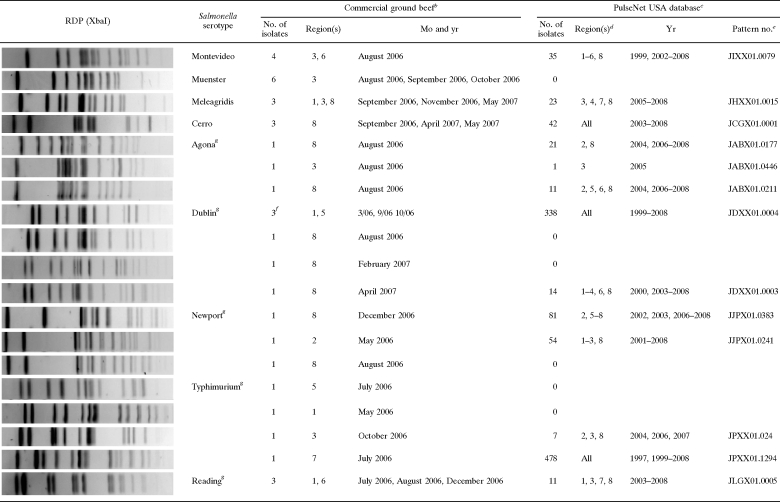
The Salmonella RDPs generated and analyzed in this study were based on PFGE separation of XbaI-, and AvrII-digested genomic DNA, using methods developed by members of PulseNet (http://www.cdc.gov/PULSENET/). In cases where DNA degradation-sensitive isolates were identified, PFGE conditions were modified to include 100 μM thiourea (25). Salmonella enterica serotype Braenderup strain H9812 was used as a control and for standardization of all gels (24). Banding patterns were analyzed and comparisons were made using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium), employing the Dice similarity coefficient in conjunction with the unweighted-pair group method using arithmetic averages for clustering. Isolates were grouped into types that likely had the same origin, based on the similarities between the RDPs. Types were defined strictly as isolates that grouped together and had indistinguishable PFGE patterns by software analysis (approximately 99.99% Dice similarity) and by visual confirmation. The unique XbaI RDPs of all drug-resistant isolates were submitted to PulseNet USA for analysis. Also, the most frequent RDP for each of the most frequent serotypes isolated (Salmonella enterica serotypes Montevideo, Muenster, Meleagridis, and Cerro) was submitted to PulseNet for analysis. The PulseNet analysis returned the pattern identification numbers for indistinguishable RDPs in their database, along with isolation dates and locations. The state health laboratory locations provided by PulseNet were placed in the corresponding BIFSCo regions used to separate the ground beef producers for comparisons (see Table 4).
TABLE 4.
Comparison of indistinguishable RDPs of selected Salmonella isolates from ground beef to RDPs in the PulseNet USA database of human Salmonella isolatesa
The Salmonella strains selected for comparison to the PulseNet database were the most frequent RDPs of Salmonella serotypes Montevideo, Muenster, Meleagridis, and Cerro and all MDR Salmonella strains resistant to four or more antibiotics. PulseNet analysis was done by Jana Austin and Peter Gerner-Smidt.
For each selected RDP from a ground beef isolate, the number of indistinguishable RDPs, the BIFSCo region, and the month and year of sample collection are provided.
Shown are the number of indistinguishable RDPs in the database, with the corresponding BIFSCo region and year of isolation/submission.
For this analysis, BIFSCo region 1 has been extended to include AK and HI.
PulseNet USA XbaI pattern accession number.
These three Salmonella serotype Dublin strains have different AvrII (or BlnI) RDPs, but for PulseNet analysis, only the XbaI RDPs that were identical were used.
MDR strains.
Statistics.
Analyses of variance for the prevalences of Salmonella in general, specific serotypes of Salmonella, and MDR Salmonella by region and sampling period were performed using GraphPad Prism software (GraphPad Software, La Jolla, CA). Specifically, the nonparametric data were analyzed using Kruskal-Wallis one-way analysis of variance by ranks with Dunn's multiple comparison posttest. P values of <0.05 were considered significantly different.
RESULTS AND DISCUSSION
The objective of this study was to characterize Salmonella present in commercial U.S. ground beef. Eighteen commercial ground beef producers participated in this study between July 2005 and June 2007. For anonymity, each producer was code numbered and located in the appropriate BIFSCo microbiological monitoring region of the United States (Fig. 1). The number of samples provided by each producer varied, and not every producer provided monthly samples. Therefore, for any given month, an average of eight establishments (range, 1 to 18 establishments) in five regions (range, one to seven regions) collected and submitted a total of 172 samples (range, 11 to 448 samples). Each establishment provided samples for an average of 11 of the 24 months of this study (range, 4 to 23 months). This resulted in a total of 4,136 samples, just over half of which were collected and analyzed in the 6 months from June through November 2006. As a result of the anomalies in the sample submission process, region 8 was somewhat overrepresented, while regions 7 and 4 were underrepresented.
Overall, Salmonella was isolated from 172 samples in our study, making the prevalence 4.2%, which is higher than that reported in other studies. The NARMS summary of Salmonella for the years 2002 through 2005 reported a Salmonella prevalence of 1%, identified by isolating 40 salmonellae in 3,904 retail ground beef samples (6, 36). In a study of retail meats in Washington, DC, only 4 of 210 ground beef samples (2%) were positive for Salmonella (35). Reports from the FSIS, during the same time period as our study, identified an approximately 2% positivity rate in their establishment-monitoring sample sets (3). The increased prevalence that we report is most likely due to our methodology. The likelihood of isolating Salmonella, if present, is increased when IMS (16) and larger sample sizes (1, 2) are used. In an interlaboratory comparison, the U.S. Meat Animal Research Center Salmonella method was shown to be significantly more sensitive at isolating Salmonella than other laboratory protocols (T. Besser, unpublished data) that did not use IMS. The larger, 65-g sample used here allows for detection of a nonuniformly distributed pathogen in the ground beef. The reports that reported lower prevalences of Salmonella did not use IMS and used a smaller (25-g) sample size.
Analysis of variance of the monthly prevalence of Salmonella showed that there was no difference between regions (Table 1). However, the prevalence of Salmonella was observed to be highest in samples collected from region 3 (6.5%; 95% confidence interval [CI], 4.4 to 8.5%) and lowest in samples collected from region 2 (1.8%; 95% CI, −0.7 to 4.4%). Because of the variable number of samples and the variable number of regions submitting samples each month, a timewise analysis of Salmonella prevalence is not presented. Such an analysis would inaccurately represent small samples obtained from one or two suppliers during some months for comparison to large sets of samples obtained from multiple sources during other months. Generally, Salmonella was more often isolated from samples collected in summer months, similar to other studies that examined seasonal prevalence in detail (7, 22, 26).
TABLE 1.
Monthly Salmonella prevalences for each region over the course of the study
| Region | Total no. of samples | No. of mosa | Avg no. of samples per mo (range)b | Mean % (95% CI)c |
|---|---|---|---|---|
| 1 | 529 | 17 | 31 (8-91) | 2.42* (−0.080 to 4.913) |
| 2 | 267 | 15 | 18 (5-55) | 1.82* (−0.723 to 4.394) |
| 3 | 691 | 23 | 30 (18-67) | 6.46* (4.383 to 8.547) |
| 5 | 461 | 12 | 38 (20-75) | 2.91* (−0.023 to 5.789) |
| 6 | 560 | 12 | 47 (22-96) | 4.14* (1.254 to 7.020) |
| 7 | 174d | 12 | 13 (1-21) | 3.28* (0.510 to 6.049) |
| 8 | 1450 | 23 | 63 (11-129) | 3.00* (0.918 to 5.083) |
The number of months out of the 24-month study in which more than two samples from a given region were received.
Average number of samples received each month.
Mean Salmonella prevalence. Values bearing common numbers of asterisks do not significantly differ (P > 0.05).
One hundred seventy-five samples were received from region 7 over 13 months, but in one month, only one sample was collected, and sample data for that month were dropped for proper analysis of prevalence.
Due to the scope of this sampling, ground beef samples were stored at −20°C after collection until processed, which in some cases was as long as 60 days. Fresh beef samples would have been preferable for Salmonella analysis because the freeze-thaw processes may have injured the Salmonella bacteria, affecting their detection, and resulted in an underestimate of levels and prevalence. However, many of the ground beef samples received were patties or products that were to be frozen before distribution to food service and commercial users, so a frozen sample was most representative in these cases. Further, controls accounting for frozen storage and thawing effects on Salmonella isolation were included with all Salmonella analyses.
The levels of Salmonella in each sample were examined by a direct plating enumeration method (data not shown). The limit of detection for this method was 2 CFU/gram of ground beef. One hundred sixty-two samples were found to have Salmonella present below this level, as indicated by their being Salmonella culture positive after enrichment and yet negative for the enumeration assay. Nine samples had Salmonella present at 2 to 4 CFU/g, and these were received from five different regions. The highest level of Salmonella observed was in one sample that had 40 CFU/g. Since the enumeration method relies on direct plating to a selective medium, the values obtained may be an underestimate, since injured cells may not grow robustly in this assay format (10).
The serotypes of all Salmonella isolates were determined (Table 2), with all isolates being Salmonella enterica subspecies enterica except one, which was supspecies IV (houtenae). Of the 28 serotypes of Salmonella isolated, Salmonella enterica serotypes Anatum, Mbandka, Montevideo, and Muenster represented 50% of the isolates. The seasonal and regional prevalences of the serotypes found were not different (P > 0.05). The most frequent serotypes were spread across four or five of the regions. Salmonella serotypes Agona, Cerro, and Typhimurium were found infrequently but also present in four or five regions. The most common serotypes reported to derive from human sources and responsible for hospitalizations in the United States are Salmonella enterica serotypes Typhimurium, Enteritidis, Heidelberg, Newport, and Javiana (15). Two of these serotypes, Salmonella serotypes Enteritidis and Javiana, were not isolated from the commercial ground beef samples. This is not unexpected, since Salmonella serotype Enteritidis is a serotype associated with poultry and eggs (13, 29), and Salmonella serotype Javiana is a serotype with poorly understood sources but is reported to occur in outbreaks associated with a variety of fresh produce items (12, 23). Compared to the 20 most frequent human serotypes reported to the CDC, only 9 of the 28 serotypes isolated from ground beef were represented. In their report of serotype profiles of analyzed pathogen reduction/hazard analysis and critical control point verification samples from ground beef, the FSIS identified serotypes similar to those that we identified here, and at approximately the same distribution (4).
TABLE 2.
Serotypes of Salmonella strains isolated from commercial ground beef (n = 173)
| Salmonella enterica serotypea | % of total | Region(s) |
|---|---|---|
| Montevideo | 21.0 | 1, 3, 5, 6, 8 |
| Anatum | 14.8 | 2, 3, 5, 6, 8 |
| Muenster | 8.5 | 3, 5, 6, 8 |
| Mbandaka | 5.7 | 1, 3, 6, 7 |
| Agona* | 5.1 | 3*, 5, 6, 7, 8* |
| Cerro | 5.1 | 1, 3, 6, 8 |
| Meleagridis | 5.1 | 1, 3, 6 |
| Typhimurium* | 4.5 | 1*, 2, 3*, 5*, 7* |
| Dublin* | 3.4 | 1*, 5*, 8* |
| Kentucky | 3.4 | 1, 3, 8 |
| Reading* | 3.4 | 1*, 3*, 6* |
| Muenchen | 3.4 | 3 |
| Thompson | 2.8 | 3, 8 |
| London | 2.3 | 6 |
| Infantis | 1.7 | 3, 7, 8 |
| Newport* | 1.7 | 2*, 8* |
| Havana | 1.7 | 3 |
| Adelaide | 0.6 | 8 |
| Benfica | 0.6 | 3 |
| Brandenburg | 0.6 | 2 |
| Heidelberg | 0.6 | 8 |
| Kiambu | 0.6 | 5 |
| Liverpool | 0.6 | 7 |
| O3,10 eh:- | 0.6 | 8 |
| Paratyphi B | 0.6 | 8 |
| SanDiego | 0.6 | 8 |
| Senftenberg | 0.6 | 5 |
| IV | 0.6 | 5 |
Serotypes with asterisks demonstrated resistance to four or more antibiotics and were isolated from samples originating in the asterisked regions.
Four samples had two serotypes of Salmonella present. These were samples from three different suppliers in different regions submitted in different months. The serotypes isolated were Salmonella enterica serotypes Anatum plus Montevideo, Anatum plus Muenchen, and Infantis plus Havana. Our protocol called for the initial serogrouping of two or three isolates from each culture plate (BGS and HEn). If the isolates were of an identical serogroup, only one was taken forward to serotyping. When different serogroups were identified in this screening, one of each serogroup was taken forward to serotyping. Therefore, it is possible that additional, multiserotype-positive samples were not identified, if both were of the same serogroup.
The antibiotic susceptibilities of each Salmonella isolate were determined. Initial screening for resistance identified 37 Salmonella isolates that grew on Tet, chloramphenicol, or ampicillin. Fourteen isolates, representing Salmonella enterica serotypes Anatum, Kentucky, London, Mbandaka, Meleagridis, Montevideo, and Reading were found to be resistant only to Tet, with MICs of 16 (n = 1), 32 (n = 6), and >32 (n = 7) μg/ml. Three isolates, each of a different serotype (Salmonella enterica serotypes Benfica, Agona, and Anatum) and from different regions (no. 3, 5, and 6) were resistant to sulfisoxazole and Tet. The remaining resistant Salmonella isolates were resistant to four or more antibiotics (Table 3), but all were found susceptible to amikacin, ciprofloxacin, and trimethoprim-sulfamethoxazole. The serotypes of these MDR Salmonella isolates were limited to Salmonella serotypes Agona, Dublin, Newport, Reading, and Typhimurium. MDR Salmonella strains were isolated from samples that came from every region, and their prevalences per region was not different (P > 0.05). The month-to-month prevalences of MDR Salmonella were also found to be not different (P > 0.05).
TABLE 3.
Serotypes and antibiotic resistance patterns of Salmonella strains resistant to four or more antibioticsa
| Salmonella serotype (no. of strains) | Result for:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | AMP | FOX | TIO | AXO | CHL | GEN | KAN | NAL | STR | FIS | TET | |
| Agona (3) | R | R | R | R | I | s | S | s | s | s | R | R |
| R | R | R | R | I | R | S | R | R | R | R | R | |
| Dublin (6) | s | s | s | s | s | R | S | s | s | R | R | R |
| s | R | s | s | s | R | S | s | s | R | R | R | |
| s | R | I | s | s | R | I | R | s | R | R | R | |
| s | R | s | s | s | R | S | R | s | R | R | R | |
| R | R | R | R | s | R | S | s | s | R | R | R | |
| R | R | R | R | I | R | S | s | s | R | R | R | |
| Newport (3) | s | s | s | s | s | R | S | R | s | R | R | R |
| R | R | R | R | I | R | S | s | s | R | R | R | |
| R | R | R | R | I | R | R | R | s | R | R | R | |
| Reading (4) | R | R | R | R | s | R | S | s | s | R | R | R |
| Typhimurium (4) | I | R | s | s | s | R | s | s | s | s | R | R |
| R | R | R | R | I | R | s | s | s | R | R | R | |
| R | R | R | R | I | R | s | R | s | R | R | R | |
R, resistant; s, sensitive; I, intermediate (with MICs increased but below the resistance breakpoint value). Drug abbreviations are as follows, with breakpoint values in parentheses: AMC, amoxicillin-clavulanic (R ≥ 32 μg/ml); AMP, ampicillin (R ≥ 32 μg/ml); FOX, cefoxitin (R ≥ 32 μg/ml); TIO, ceftiofur (R ≥ 8 μg/ml); AXO, ceftriaxone (s ≤ 8 μg/ml, I = 16 to 32 μg/ml); CHL, chloramphenicol (R ≥ 32 μg/ml); GEN, gentamicin (R ≥ 16 μg/ml, I = 8 μg/ml); KAN, kanamycin (R ≥ 64 μg/ml); NAL, nalidixic acid (R ≥ 32 μg/ml); STR, streptomycin (R ≥ 64 μg/ml); FIS, sulfisoxazol (R ≥ 256 μg/ml); and TET, tetracycline (R ≥ 32 μg/ml).
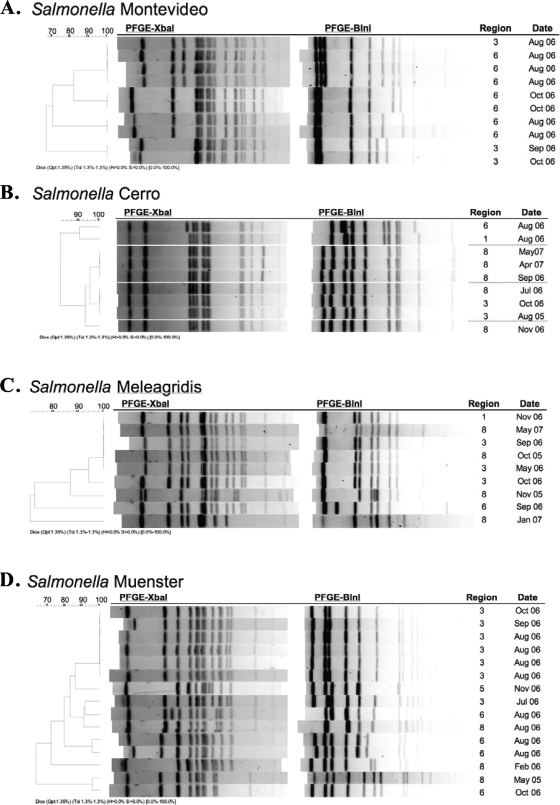
In this study, all of the Salmonella isolates were subjected to PFGE using two enzymes, XbaI and AvrII. The two-enzyme analysis of the PFGE RDPs showed that the isolates clustered by serotype (data not shown). The analysis further showed that there were a number of instances where isolates of indistinguishable patterns were present. For instance, Salmonella serotype Montevideo, the serotype most frequently isolated, was found to have 31 RDPs. As such, there were four instances where the PFGE patterns of two or more Salmonella serotype Montevideo isolates were indistinguishable (Fig. 2A). In three of these cases, though, the indistinguishable RDPs were potentially related because they were of isolates found in samples collected within days of one another at the same establishment. Further examination of the RDPs offers a number of examples where unrelated, indistinguishable isolates were found in multiple regions and/or at different sample collection times (Fig. 2B to D). Six Salmonella serotype Cerro isolates had two very similar RDPs (three each) that were from samples collected by four establishments in regions 3 and 8 over a period of 14 months. Indistinguishable isolates of Salmonella serotype Meleagridis were identified in three noncontiguous regions (no. 1, 3, and 8) over the course of 20 months (October 2005 to May 2007). Six isolates of Salmonella serotype Muenster had one RDP. While three of these isolates were from samples collected by one establishment in region 3 during the month of August, the other three were from a second establishment in region 3 and collected in three separate months. When the PFGE RDPs of antibiotic-resistant Salmonella isolates were examined, only three of the four MDR Salmonella serotype Reading strains had indistinguishable patterns. These three isolates were each from samples submitted by different establishments during different collection periods. Three of the Salmonella serotype Dublin isolates that were resistant to multiple antibiotics had indistinguishable XbaI RDPs but different AvrII RDPs.
FIG. 2.
Analysis of XbaI and AvrII PFGE patterns (AvrII patterns are analyzed as the isoschizomer BlnI) of Salmonella serotypes exhibiting indistinguishable RDPs, with BIFSCo microbiological monitoring regions and month and year of sample collection provided.
To better understand the roles these RDPs would have at linking the isolates from ground beef to other occurrences, the most common XbaI RDPs of Salmonella serotypes Montevideo, Muenster, Meleagridis, and Cerro were compared to RDPs in the PulseNet USA Salmonella database (Table 4). The most common Salmonella serotype Muenster RDP was not present in the PulseNet database. The most common RDPs of the other three serotypes, Salmonella serotypes Montevideo, Meleagridis, and Cerro, were present at over 20 times each and in four or more regions over periods longer than 4 years. The wide regional distribution of indistinguishable RDPs of ground beef isolates and PulseNet isolates may lead one to speculate that the beef trim used over time and across regions or that the cattle sources of the materials were potentially shared by different ground beef producers. However, given the nature of ground beef production, with some producers generating 30,000 pounds per hour, it is highly unlikely that materials from the same source would be widely spread temporally or geographically.
MDR Salmonella isolates have been suggested to be more virulent than non-MDR Salmonella isolates (19, 20), so we hypothesized that the MDR Salmonella isolates from ground beef would all be indistinguishable from one or more isolates in the PulseNet database. However, when we compared the XbaI RDPs of all the isolates with resistance to four or more antibiotics to the XbaI RDPs in the PulseNet database, we found that approximately one-quarter (26%) were not present. When indistinguishable RDPs were identified, they were distributed over various regions and many years. The XbaI RDP of one MDR Salmonella serotype Typhimurium isolate from ground beef had 478 matches in the PulseNet database. Only one of the MDR Salmonella RDPs matched any outbreak-related isolates in PulseNet, and these were three Salmonella serotype Dublin isolates. However, these three Salmonella serotype Dublin isolates were distinguished from one another by different AvrII patterns that are not available in the PulseNet database.
Although two strains of Salmonella may have indistinguishable RDPs after digestion with two restriction enzymes, it does not necessarily follow that those strains are identical or even related. It has been suggested that PFGE analysis based on DNA digestion patterns of up to six enzymes is required for an accurate estimation of strain relatedness in organisms like Escherichia coli (17). Other researchers have reported that for source-tracking studies related to food-borne illness outbreaks, one enzyme PFGE analysis is useful only if supported by reliable epidemiological data (8, 32). The data presented here show that strains with matching RDPs, even after digestion with two restriction enzymes, can originate from multiple ground beef sources and that those sources may be separated by large distances.
In conclusion, this study analyzed 4,136 commercial ground beef samples for the presence of Salmonella in samples collected from seven regions of the United States over a period of 24 months. The overall prevalence of Salmonella was 4.2%, and the prevalence of MDR Salmonella was 0.6%. Salmonella serotypes Montevideo, Anatum, Muenster, and Mbandaka comprised half of the isolates. The most common MDR serotypes were Salmonella serotypes Dublin, Reading, and Typhimurium. PFGE analysis showed that indistinguishable two-enzyme RDPs could be observed in isolates of the same serotype from different regions and different time periods and that indistinguishable XbaI RDPs were also present in the PulseNet database. The data suggest that attempts to identify sources contributing to Salmonella in ground beef based on serotype, antibiogram, and PFGE pattern cannot be made in the absence of additional supporting documentation or evidence.
Acknowledgments
We gratefully thank the participating ground beef producers for their role in supplying, collecting, and submitting samples for this work. We also thank Dennis Johnson for assistance in organizing the receipt and maintenance of confidentiality of suppliers. PulseNet USA data were kindly provided through Jana Austin and Peter Gerner-Smidt. We thank Greg Smith, Bruce Jasch, Frank Reno, Emily Griese, and Sara Schumacher for technical support; Debbie Kummer for secretarial support; and Terrance Arthur and Dayna Harhay for scientific support and critical review of the manuscript.
The use of product names is necessary for factual reporting on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by the USDA does not imply approval of the product to the exclusion of others that may also be suitable.
Footnotes
Published ahead of print on 6 February 2009.
REFERENCES
- 1.Andrews, W. H., and T. Hammack. 2008. Chapter 5, Salmonella. In Bacteriological analytical manual, 8th ed., revision A, 1998. U.S. Food and Drug Administration, Silver Spring, MD. http://www.cfsan.fda.gov/∼ebam/bam-5.html. Accessed 21 January 2009.
- 2.Anonymous. 2008. Microbiology laboratory guidebook. U.S. Department of Agriculture, Washington, DC. http://www.fsis.usda.gov/Science/Microbiological_Lab_Guidebook/. Accessed 5 November 2008.
- 3.Anonymous. 2008. Progress report on Salmonella testing of raw meat and poultry products, 1998-2007. U.S. Department of Agriculture, Washington, DC. http://www.fsis.usda.gov/science/progress_report_salmonella_testing/index.asp. Accessed 5 November 2008.
- 4.Anonymous. 2007. Serotypes profile of Salmonella isolates from meat and poultry products: January 1998 through December 2006. U.S. Department of Agriculture, Washington, DC. http://www.fsis.usda.gov/Science/Serotypes_Profile_Salmonella_Isolates/index.asp. Accessed 5 November 2008.
- 5.Anonymous. 2008. Bad bug book: foodborne pathogenic microorganisms and natural toxins handbook. U.S. Food and Drug Administration, Silver Spring, MD. http://www.cfsan.fda.gov/∼mow/chap1.html. Accessed 5 November 2008.
- 6.Anonymous. 2008. NARMS retail meat annual report, 2005. U.S. Food and Drug Administration, Washington, DC. http://www.fda.gov/cvm/2005NARMSAnnualRpt.htm. Accessed 5 November 2008.
- 7.Barkocy-Gallagher, G. A., T. M. Arthur, M. Rivera-Betancourt, X. Nou, S. D. Shackelford, T. L. Wheeler, and M. Koohmaraie. 2003. Seasonal prevalence of Shiga toxin-producing Escherichia coli, including O157:H7 and non-O157 serotypes, and Salmonella in commercial beef processing plants. J. Food Prot. 66:1978-1986. [DOI] [PubMed] [Google Scholar]
- 8.Barrett, T. J., P. Gerner-Smidt, and B. Swaminathan. 2006. Interpretation of pulsed-field gel electrophoresis patterns in foodborne disease investigations and surveillance. Foodborne Pathog. Dis. 3:20-31. [DOI] [PubMed] [Google Scholar]
- 9.Berge, A. C. B., E. L. Dueger, and W. M. Sischo. 2006. Comparison of Salmonella enterica serovar distribution and antibiotic resistance patterns in wastewater at municipal water treatment plants in two California cities. J. Appl. Microbiol. 101:1309-1315. [DOI] [PubMed] [Google Scholar]
- 10.Brichta-Harhay, D. M., T. M. Arthur, J. M. Bosilevac, M. N. Guerini, N. Kalchayanand, and M. Koohmaraie. 2007. Enumeration of Salmonella and Escherichia coli O157:H7 in ground beef, cattle carcass, hide and faecal samples using direct plating methods. J. Appl. Microbiol. 103:1657-1668. [DOI] [PubMed] [Google Scholar]
- 11.CDC. 2006. Multistate outbreak of Salmonella Typhimurium infections associated with eating ground beef—United States, 2004. MMWR Morb. Mortal. Wkly. Rep. 55:180-182. [PubMed] [Google Scholar]
- 12.CDC. 2005. Outbreaks of Salmonella infections associated with eating Roma tomatoes—United States and Canada, 2004. MMWR Morb. Mortal. Wkly. Rep. 54:325-328. [PubMed] [Google Scholar]
- 13.CDC. 2003. Outbreaks of Salmonella serotype Enteritidis infection associated with eating shell eggs—United States, 1999-2001. MMWR Morb. Mortal. Wkly. Rep. 51:1149-1152. [PubMed] [Google Scholar]
- 14.CDC. 2002. Outbreak of multidrug-resistant Salmonella Newport—United States, January-April 2002. MMWR Morb. Mortal. Wkly. Rep. 51:545-548. [PubMed] [Google Scholar]
- 15.CDC. 2007. Salmonella surveillance: annual summary, 2005. U.S. Department of Health and Human Services, CDC, Atlanta, GA. http://www.cdc.gov/ncidod/DBMD/phlisdata/salmtab/2005/SalmonellaIntroduction2005.pdf.
- 16.Cudjoe, K. S., and R. Krona. 1997. Detection of Salmonella from raw food samples using Dynabeads anti-Salmnella and a conventional reference method. Int. J. Food Mircrobiol. 37:55-62. [DOI] [PubMed] [Google Scholar]
- 17.Davis, M. A., D. D. Hancock, T. E. Besser, D. H. Rice, C. J. Hovde, R. Digiacomo, M. Samadpour, and D. R. Call. 2003. Correlation between geographic distance and genetic similarity in an international collection of bovine faecal Escherichia coli O157:H7 isolates. Epidemiol. Infect. 131:923-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eblen, D. R., K. E. Barlo, and A. L. Naugle. 2006. U.S. Food Safety and Inspection Service testing for Salmonella in selected raw meat and poultry products in the United States, 1998 through 2003: an establishment-level analysis. J. Food Prot. 69:2600-2606. [DOI] [PubMed] [Google Scholar]
- 19.Fluit, A. D. 2005. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol. Med. Microbiol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 20.Foley, S. L., and A. M. Lynne. 2008. Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J. Anim. Sci. 86:E173-E187. [DOI] [PubMed] [Google Scholar]
- 21.Gatto, A. J., T. M. Peters, J. Green, I. S. Fisher, O. N. Gill, S. J. O'Brien, C. Maguire, C. Berghold, I. Lederer, P. Gerner-Smidt, M. Torpdahl, A. Siitonen, S. Lukinmaa, H. Tschäpe, R. Prager, I. Luzzi, A. M. Dionisi, W. K. van der Zwaluw, M. Heck, J. Coia, D. Brown, M. Usera, A. Echeita, and E. J. Threlfall. 2006. Distribution of molecular subtypes within Salmonella enterica serotype Enteritidis phage type 4 and S. Typhimurium definitive phage type 104 in nine European countries, 2000-2004: results of an international multi-centre study. Epidemiol. Infect. 134:729-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hald, T., and J. S. Andersen. 2001. Trends and seasonal variations in the occurrence of Salmonella in pigs, pork and humans in Denmark, 1995-2000. Berl. Munch. Tierarztl. Wochenschr. 114:346-349. [PubMed] [Google Scholar]
- 23.Hedberg, C. W., F. J. Angulo, K. E. White, C. W. Langkop, W. L. Schell, M. G. Stobierski, A. Schuchat, J. M. Besser, S. Dietrich, L. Helsel, P. M. Griffin, J. W. McFarland, and M. T. Osterholm. 1999. Outbreaks of salmonellosis associated with eating uncooked tomatoes: implications for public health. Epidemiol. Infect. 122:385-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter, S. B., P. Vauterin, M. A. Lambert-Fair, M. S. Van Duyne, K. Kubota, L. Graves, D. Wrigley, T. Barrett, and E. Ribot. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liesegang, A., and H. Tschäpe, H. 2002. Modified pulsed-field gel electrophoresis method for DNA degradation-sensitive Salmonella enterica and Escherichia coli strains. Int. J. Med. Microbiol. 291:645-648. [DOI] [PubMed] [Google Scholar]
- 26.Maharjan, M., V. Joshi, D. D. Joshi, and P. Manandhar. 2006. Prevalence of Salmonella species in various raw meat samples of a local market in Kathmandu. Ann. N. Y. Acad. Sci. 1081:249-256. [DOI] [PubMed] [Google Scholar]
- 27.Malorny, B., J. Hoorfar, C. Bunge, and R. Helmuth. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishu, B., J. Koehler, L. A. Lee, Rodrigue, F. H. Brenner, P. Blake, and R. V. Tauxe. 1994. Outbreaks of Salmonella enteriditis infections in the United States, 1985-1991. J. Infect. Dis. 169:547-5211. [DOI] [PubMed] [Google Scholar]
- 30.Nucera, D. M., C. W. Maddox, P. Hoien-Dalen, and R. M. Weigel. 2006. Comparison of API 20E and invA PCR for identification of Salmonella enterica isolates from swine production units. J. Clin. Microbiol. 44:3388-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahn, K., S. A. De Grandis, R. C. Clarke, S. A. McEwen, J. E. Galan, C. Ginocchio, R. Curtiss, and C. L. Gyles. 1992. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6:271-279. [DOI] [PubMed] [Google Scholar]
- 32.Singer, R. S., W. M. Sischo, and T. E. Carpenter. 2004. Exploration of biases that affect the interpretation of restriction fragment patterns produced by pulsed-field gel electrophoresis. J. Clin. Microbiol. 42:5502-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Usera, M. A., A. Aladueña, R. González, M. De La Fuente. A. J. G.-P. Frías, and M. A. Echeita. 2002. Antibiotic resistance of Salmonella spp. from animal sources in Spain in 1996 and 2000. J. Food Prot. 65:768-773. [DOI] [PubMed] [Google Scholar]
- 34.White, D. G., S. Zhao, R. Sudler, S. Ayers, S. Friedman, S. Chen, P. F. McDermott, S. McDermott, D. D. Wagner, and J. Meng. 2001. The isolation of antibiotic-resistant salmonella from retail ground meats. N. Engl. J. Med. 345:1147-1154. [DOI] [PubMed] [Google Scholar]
- 35.Zhao, C., B. Ge, J. De Villena, R. Sudler, E. Yeh, S. Zhao, D. G. White, D. Wagner, and J. Meng. 2001. Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the greater Washington, D.C., area. Appl. Environ. Microbiol. 67:5431-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, S., P. F. McDermott, S. Friedman, J. Abbott, S. Ayers, A. Glenn, E. Hall-Robinson, S. K. Hubert, H. Harbottle, R. D. Walke, T. M. Chiller, and D. G. White. 2006. Antimicrobial resistance and genetic relatedness among Salmonella from retail foods of animal origin: NARMS retail meat surveillance. Foodborne Pathog. Dis. 3:106-117. [DOI] [PubMed] [Google Scholar]
- 37.Zhao, T., M. P. Doyle, P. J. Fedorka-Cray, P. Zhao, and S. Ladely. 2002. Occurrence of Salmonella enterica serotype Typhimurium DT104A in retail ground beef. J. Food Prot. 65:403-407. [DOI] [PubMed] [Google Scholar]
- 38.Zozo, N., O. Vandenberg, B. Bisimwa, A. Dediste, P. Donnen, G. Zissis, and J. Butzler. 2004. Serotypes and antibiotic resistance patterns of Salmonella species isolates in the province of South-Kivu, Democratic Republic of Congo. Abstr. 14th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. P772. Clin. Microbiol. Infect. 10(Suppl. 3):194. [Google Scholar]