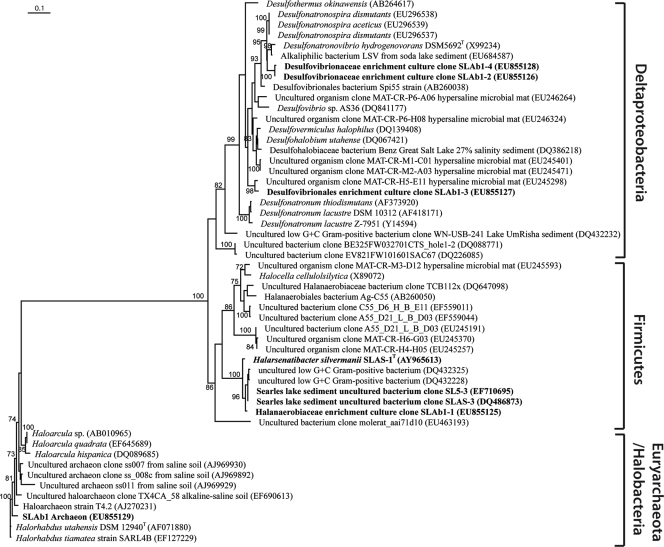
FIG. 4.
Phylogenetic analysis of Halarsenatibacter silvermanii strain SLAS-1T and the antibiotic enrichment culture, showing the relationship of the 16S rRNA genes of isolates and enrichment culture to known species and environmental clones. Sequences from this study are shown in boldface, and the GenBank accession numbers for sequences obtained from the NCBI database are shown in parentheses. All sequences were automatically aligned according to the SILVA SSU reference alignment (SILVA Incremental Aligner [43]). All aligned sequences were imported and analyzed using the ARB software package (version 08.08.13) (28). Manual refinement of the alignment was carried out in ARB editor4, taking into account the secondary structure information of the rRNA. Tree reconstruction was performed with 52 sequences using maximum likelihood (Rhyml version 2.4.5) (11). The final tree was calculated with 503 positions (all ambiguous bases and nonaligned positions were omitted from the analysis) using PHYML (model, GTR and GAMMA). Bootstrap values were generated by 100 replications, and bootstrap values above 70 are shown next to the branches. The bar in the upper left corner refers to 0.1 fixed nucleotide substitutions per site.