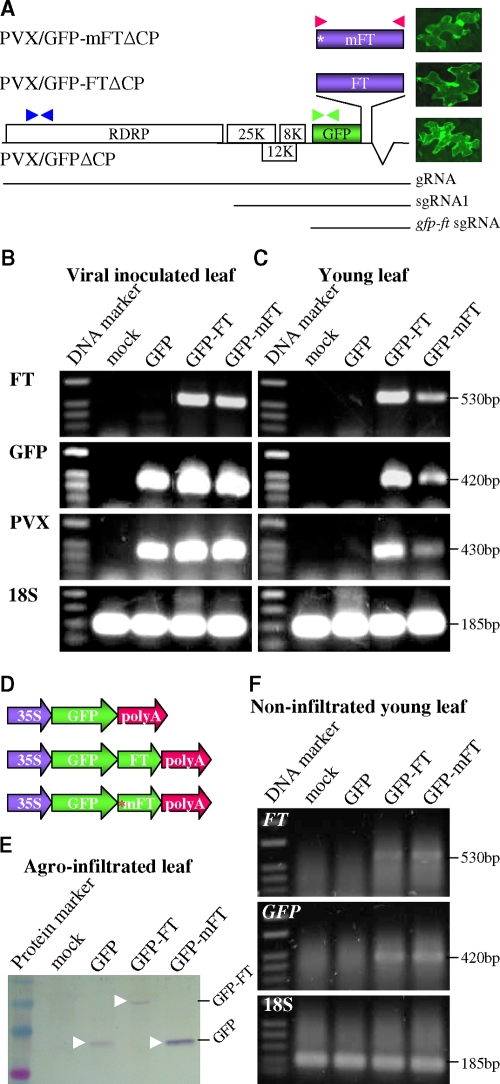
FIG. 1.
(A) PVX-based RMA vectors. Viral 7.0-kb genomic gRNA, 2.6-kb subgenomic sgRNA1, and 1.4-kb GFP-FT sgRNA are indicated. The CP gene was deleted. The positions of a stop codon (*) replacing FT start codon in PVX/GFP-mFTΔCP, and three sets of RT-PCR primers for the detection of FT (▸◂ [red], PP354/PP356), GFP (▸◂ [green], PP371/PP372), or PVX (▸◂ [blue], PP269/PP373) RNAs are indicated. Individual epidermal cells expressing free GFP or GFP-FT fusion protein showed green fluorescence. (B and C) RT-PCR analysis of FT, GFP, and PVX RNA and 18S rRNA in inoculated and newly growing young leaves, including shoot apices of N. benthamiana. RNAs were extracted from leaves of plants mock inoculated (mock) or inoculated with PVX/GFPΔCP (GFP), PVX/GFP-FTΔCP (GFP-FT), or PVX/GFP-mFTΔCP (GFP-mFT). The 1-kb DNA ladder and sizes of RT-PCR products are indicated. (D) Construction of 35S promoter-controlled FT gene expression cassettes for a transient agroinfiltration assay of RNA mobility. The position of an introduced stop codon (*) replacing the FT gene start codon in 35S-GFP-mFT-poly(A) is indicated. (E) Expression of free GFP or GFP-FT fusion protein. Total proteins were extracted from N. benthamiana leaves infiltrated with agrobacteria carrying the 35S-controlled gene expression cassette and were analyzed by Western blotting with a GFP-specific antibody. The positions of free GFP and GFP-FT fusion protein are indicated. (F) FT and GFP mRNAs in nonagroinfiltrated newly developed systemic young leaves were analyzed by RT-PCR. Samples are from plants with mock infiltration (mock) or infiltrated with agrobacteria carrying 35S-GFP-poly(A) (GFP), 35S-GFP-FT-poly(A) (GFP-FT), or 35S-GFP-mFT-poly(A) (GFP-mFT). The sizes of RT-PCR products, free and GFP-FT fusion proteins, and the prestained protein markers or 1-kb DNA ladder are indicated.