FIG. 8.
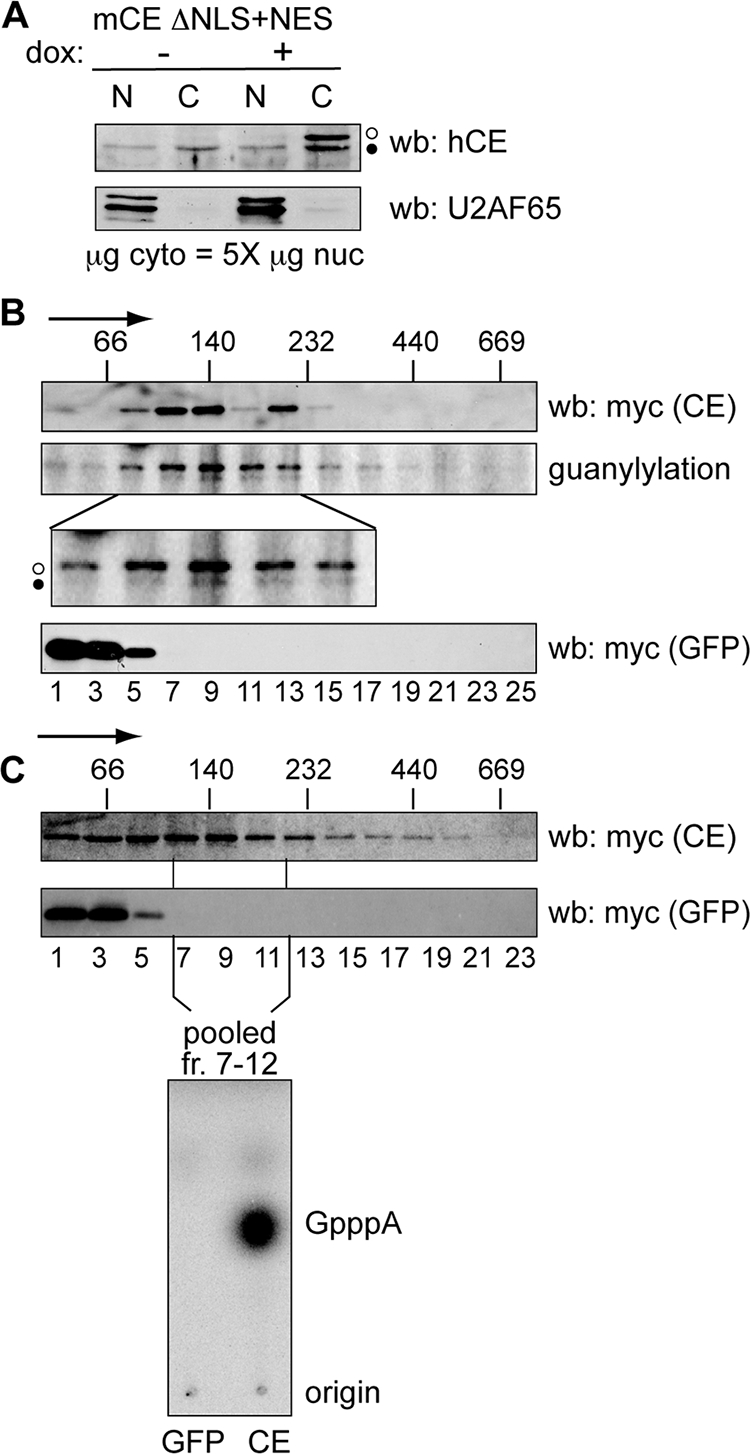
Identification of a cytoplasmic capping enzyme complex. (A) U2OS cells stably expressing mCE ΔNLS+NES were cultured for 24 h in the absence (−) or presence (+) of doxycycline (dox), and nuclear (N) and a fivefold-greater amount of cytoplasmic (C) protein was analyzed by Western blotting (wb) with affinity-purified capping enzyme antibody (top) or antibody to U2AF65 (bottom). The open circles correspond to recombinant protein and the filled circles correspond to endogenous capping enzyme. (B) Doxycycline was added to induce recombinant protein expression in cell lines stably transfected with tetracycline-inducible forms of mCE ΔNLS+NES or GFP. Cells were treated with 0.25% formaldehyde for 15 min prior to lysis, and cytoplasmic extracts were separated on linear 10 to 40% glycerol gradients. The sedimentation of mCE ΔNLS+NES or GFP was determined by Western blotting of individual fractions with antibody to the myc tag on each of these proteins, and the cosedimentation of mCE ΔNLS+NES (middle, open circle) with endogenous capping enzyme (CE) (filled circle) was determined by guanylylation assay. The lower panel is an enlargement of that portion of the gradient containing the 140-kDa capping enzyme complex. The positions of MW standards that were run in parallel gradients are indicated on the top panel, with the arrow indicating the direction of sedimentation. (C) mCE ΔNLS+NES and GFP were induced as in panel B by addition of tetracycline to lines of U2OS cells expressing each of these proteins. Cytoplasmic extracts were separated on 10 to 40% glycerol gradients, and capping enzyme and GFP were identified by Western blotting of individual fractions using antibody to the myc tag on each of these proteins. The fractions corresponding to the 140-kDa complex from panel B were pooled and recovered using beads containing immobilized anti-myc monoclonal antibody. These were incubated with [α-32P]GTP, ATP, and 5′-P-RNA, and RNA recovered from this reaction was digested with P1 nuclease and separated on PEI-cellulose TLC to identify the product of the capping reaction.
