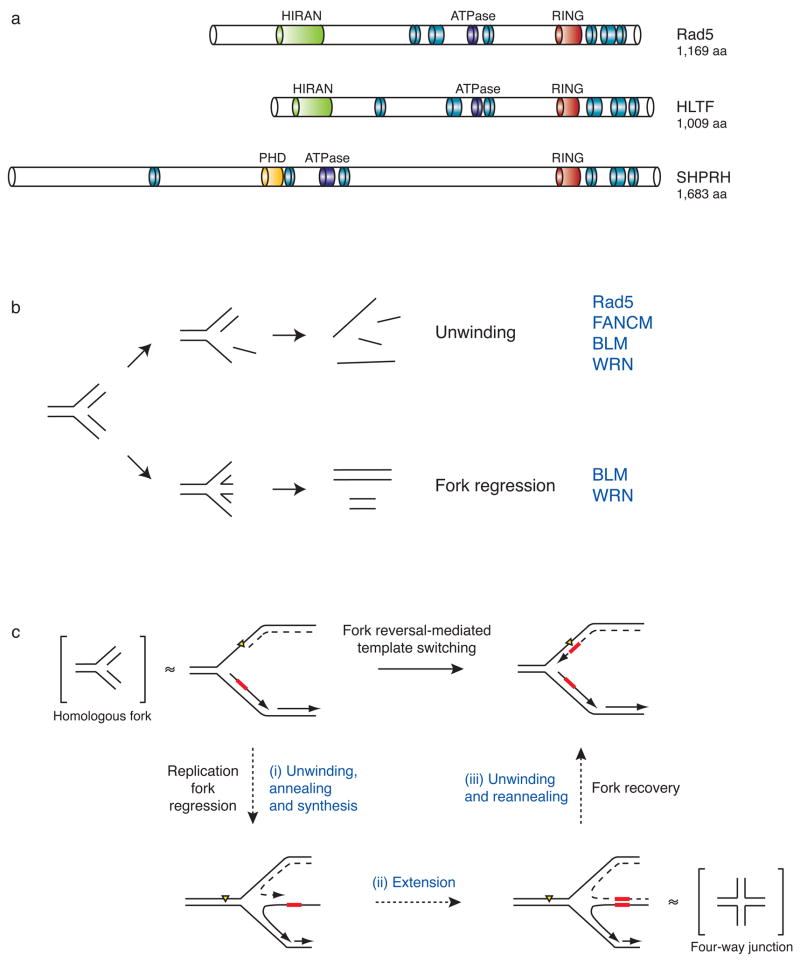
Figure 4. Domain architecture and functions of the E3 ubiquitin ligase Rad5 and its putative orthologs SHPRH and HLTF.
(a) Structural comparisons between S. cerevisiae Rad5 and its putative human orthologs, HLTF and SHPRH. Blue and purple modules represent the seven helicase motifs characteristic of the SWI/SNF2 family of ATP-driven motor proteins. Although these motifs are spread over the length of the protein, they are collectively referred to as a helicase domain. Other domain names and descriptions are as listed through the NCBI conserved domain database and are described in the text.
(b) The possible effects of helicase activity on a model homologous fork substrate. Unwinding and annealing of the nascent and parental DNA strands (bottom) results in fork regression and double-stranded DNA products, while unwinding alone (top) leads to single-stranded DNA products. Helicases known to exhibit these activities are listed in blue.
(c) A possible mechanism for template switching mediated by fork reversal. Fork regression requires (i) concerted unwinding and annealing of the newly synthesized DNA strands, (ii) extension of the strand formed by the stalled polymerase past the sequence where the lesion is found on the parental template, and (iii) unwinding of the newly formed duplex so that the nascent strands can reanneal to their original templates and restore the fork to its proper conformation. Model four-way junction and homologous fork structures, which are known substrates of Rad5 in vitro62, are placed in brackets adjacent to the fork structures they are thought to mimic. Yellow triangle = replication-blocking lesion. Dashed line = leading strand. Red box = template.