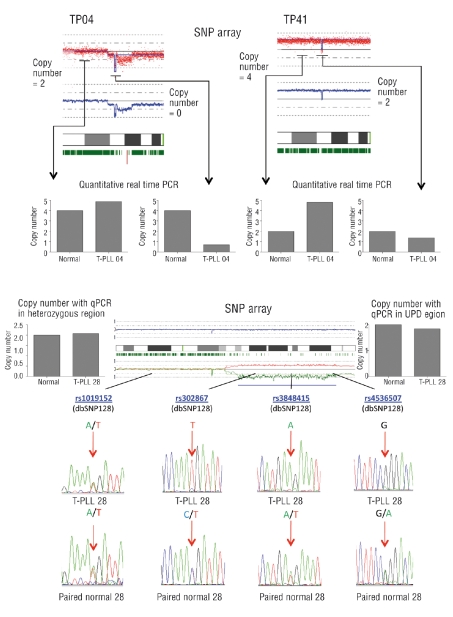
Figure 3.
Validation of copy number analysis and acquired UPD. (A) Copy number results obtained by SNP arrays were validated by performing quantitative real-time PCR on genomic DNA of samples containing putative deletions of the FOXP1 gene. Chromosome views of chromosome 3 are displayed. In sample TP04, FOXP1 was putatively homozygously deleted (left image, FOXP1 locus = copy number 0, adjacent regions = copy number: two), while in sample TP41, FOXP1 was deleted heterozygously in a tetraploid setting (right image, FOXP1 locus = copy number: two, adjacent regions = copy number: four). Quantitative real-time PCR in the corresponding T-PLL samples and their matched normal DNA confirmed the copy number states estimated by the allelokaryotyping software. (B) Acquired UPD detected by SNP arrays was validated using sample TP28 on chromosome 17. The chromosome view of chromosome 17 in sample TP28 is depicted. Acquired UPD is present on chromosome 17q, as visualized by the divergence of the estimated allele specific copy number (red and green lines in the lower panel of the image), indicating the duplication of one allele and concomitant loss of the other allele and loss of heterozygosity, as evidenced by the abrupt absence of heterozygous SNP calls (vertical green bars directly below the cytoband image). Chromatographs of sequenced SNPs within the acquired UPD region and the adjacent heterozygous region in T-PLL DNA (TP28) and matched normal DNA (paired normal DNA) show that SNPs were homozygous in the T-PLL sample in the acquired UPD region and heterozygous in the matched normal DNA. In the adjacent region, which did not display acquired UPD, the sequenced SNPs were heterozygous in both T-PLL and matched normal samples. Quantitative real-time PCR confirmed a copy number state of two in all regions sequenced in each of the samples.