Abstract
Sterols and sphingolipids are limited to eukaryotic cells, and their interaction has been proposed to favor formation of lipid microdomains. Although there is abundant biophysical evidence demonstrating their interaction in simple systems, convincing evidence is lacking to show that they function together in cells. Using lipid analysis by mass spectrometry and a genetic approach on mutants in sterol metabolism, we show that cells adjust their membrane composition in response to mutant sterol structures preferentially by changing their sphingolipid composition. Systematic combination of mutations in sterol biosynthesis with mutants in sphingolipid hydroxylation and head group turnover give a large number of synthetic and suppression phenotypes. Our unbiased approach provides compelling evidence that sterols and sphingolipids function together in cells. We were not able to correlate any cellular phenotype we measured with plasma membrane fluidity as measured using fluorescence anisotropy. This questions whether the increase in liquid order phases that can be induced by sterol–sphingolipid interactions plays an important role in cells. Our data revealing that cells have a mechanism to sense the quality of their membrane sterol composition has led us to suggest that proteins might recognize sterol–sphingolipid complexes and to hypothesize the coevolution of sterols and sphingolipids.
INTRODUCTION
Sterols are major plasma membrane components of most eukaryotic membranes, although their precise structure differs between the kingdoms; animals contain cholesterol, and plants have sitosterol, campesterol, and stigmasterol, whereas fungi use mainly ergosterol. The plant and yeast sterols both differ from cholesterol in their side chain, and yeast sterols also have an extra double bond in the B ring. As sterols appear only in eukaryotes, it is likely that they confer properties to membranes or provide functions that may not be required in prokaryotes and possibly not even in all eukaryotes (Matyash et al., 2004). Cholesterol has been shown to modulate membrane thickness in artificial membranes, and this property has been proposed to play a role in membrane protein localization in vivo (Bretscher and Munro, 1993) although an alternative explanation for the control of membrane thickness has been postulated (Mitra et al., 2004). Recently it has been shown that proteins and lipids do not freely diffuse over the entire surface of the cell, and it has been proposed that eukaryotic plasma membranes contain micro/nanodomains (for review see Simons and Ikonen, 1997; Kusumi et al., 2005; Jacobson et al., 2007). Indeed, different hypotheses exist to explain domain formation. The raft hypothesis has two basic tenets (Simons and Ikonen, 1997): 1) that sterols and sphingolipids interact specifically in biological membranes and 2) that their interaction causes an increase in membrane lipid order, creating membrane microdomains that affect protein function, diffusion, and/or localization. To our knowledge, there is currently a lack of evidence to demonstrate a specific role of lipid order in biological membranes as postulated by the raft hypothesis. Another hypothesis, known as the lipid shell hypothesis, suggests that domains are formed through association of specific lipids, in particular sterols and sphingolipids, with proteins (Anderson and Jacobson, 2002).
There is clear biophysical evidence that sterols and sphingolipids can segregate from other lipids in simple artificial membrane systems to form liquid ordered domains (Ahmed et al., 1997). Sterol partitioning experiments between membranes in vitro also suggest that they have affinity for membranes with a high content of sphingolipids (Wattenberg and Silbert, 1983). Sterols have been shown to have a condensing effect on artificial membranes (Radhakrishnan and McConnell, 2005), and sterol-sphingomyelin–condensed complexes have been characterized (Radhakrishnan et al., 2001). Some evidence exists in yeast suggesting a genetic interaction between mutants in sterol and sphingolipid biosynthesis (Baudry et al., 2001; Eisenkolb et al., 2002; Jin et al., 2008); however, little, if any, convincing evidence exists to show that these two lipid species function together in complex biological membranes. Therefore, the basic question of whether sterols and sphingolipids interact functionally and preferentially in biological systems remains unsolved and is a major focus of this study.
Nevertheless, various lines of evidence suggest a possible link between these two lipid classes. Sterols and sphingolipids are concomitantly affected in certain diseases. In Niemann Pick disease, although the primary defect is not yet completely certain, defects in sphingolipid and cholesterol trafficking appear to be interdependent (Puri et al., 1999; Pagano et al., 2000; Vance, 2006). One of the proposed functions of amyloid beta and presenilin is in control of sphingomyelin and cholesterol amounts in the brain (Grimm et al., 2005), which could affect the ontology of Alzheimer's. Sphingolipid depletion has also been shown to influence the SREBP pathway, controlling cholesterol and lipid biosynthesis (Scheek et al., 1997).
In this study, we combine quantitative lipid analysis and genetics in an unbiased approach to study yeast mutants in sterol synthesis. This systematic analysis provides evidence that yeast cells have a mechanism to respond to the presence of specific sterol structures in their membranes and adjust their sphingolipid composition accordingly. The genetic analysis provides convincing evidence that sterols and sphingolipids function together in cells to carry out multiple functions. Furthermore, our results suggest that sterol and sphingolipid structures have coevolved to provide optimal interaction properties between the two lipid species. Our results support the assertion of the raft hypothesis that sterols and sphingolipids interact preferentially in biological membranes; however, our initial studies of plasma membrane fluidity suggest that the increased order that can be induced by sterol–sphingolipid interaction is not the most important factor for any of the several physiological readouts we have investigated.
MATERIALS AND METHODS
Strain Construction, Growth, Plating Assays, Protein Extraction, and Western Blotting
All mutations were constructed using standard gene disruption procedures, with complete removal of open reading frames, in our strain background. Double mutants were obtained by genetic crosses. Strains are listed in Supplementary Table S1. Cerulean (cyan fluorescent protein [CFP]; Rizzo et al., 2004) was used to fluorescently tag Pdr12p in the genome using plasmid pBS10 (Yeast Resource Center, University of Washington). Yeast were grown in rich medium (1% yeast extract, 2% peptone, 2% glucose, and 40 mg/l uracil and adenine) to log phase for lipidomic and sterol analysis. Forty OD600 units of cells were harvested by centrifugation, washed with water, and frozen. For the plating assays, yeast were grown to stationary phase and diluted to 107 cells per ml in water, and 10-fold serial dilutions were pinned onto agar plates containing rich medium with additives adjusted to pH 5.5 (40 mM MES) unless indicated. For Western blot analyses, proteins were extracted after trichloroacetic acid denaturation as previously described (Urban et al., 2007). Phospho-Ypk1 and phospho-Sch9 were detected using an anti-serum described in Wanke et al. (2008). Anti-Ypk1 antiserum was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Lipid Standards
All lipid standards were obtained from Avanti Polar Lipids (Alabaster, AL), with the exception of C19:0-ceramide and dioctanoyl glycerophosphoinositol (GPIns), which were obtained from Matreya (Pleasant Gap, PA) and Echelon Biosciences (Salt Lake City, UT), respectively.
Lipid Analysis by Electrospray Ionization–Mass Spectrometry
Lipids were extracted from 40 OD600-equivalents of cells and as described (Guan and Wenk, 2006), with the following modifications. Five micrograms of dimyristoyl glycerophosphocholine (GPCho), 20 μg of dimyristoyl glycerophosphoethanolamine (GPEtn), 4 μg of dioctyl GPIns, 15 μg of didocosahexaenoyl glycerophosphoserine (GPSer), and 3 μg of C19:0-ceramide were added as internal standards. Quantification of individual molecular species was carried out using multiple reaction monitoring (MRM) with a 4000 Q-Trap mass spectrometer (Applied Biosystems, Foster City, CA). Typically 25 μl of samples was injected for analysis. The inlet system consisted of an auto-sampler and a pump, and chloroform–methanol (1:1, vol/vol) at a flow rate of 200 μl/min was used as the mobile phase. In these experiments, the first quadrupole, Q1, was set to pass the precursor ion of interest to the collision cell, Q2, where it underwent collision-induced dissociation. The third quadruple, Q3, was set to pass the structure-specific product ion characteristic of the precursor lipid of interest. Each individual ion dissociation pathway was optimized with regard to collision energy to minimize variations in relative ion abundance due to differences in rates of dissociation (Guan and Wenk, 2006). Lipid levels were calculated relative to relevant internal standards. Comparison of the means of wild-type and individual genotypes from three independent experiments were performed. The quantities of lipids are expressed as ion intensities relative to wild-type levels, converted to a log10 scale. The difference in levels of individual lipid species between wild-type and individual genotypes was determined statistically using the Kruskal-Wallis test.
Sterol Analysis by Gas Liquid Chromatography–Mass Spectrometry
Four micrograms of cholesterol standard were added to 40 A600 equivalents of washed cells, and total sterols were extracted three times with petrol ether after alkaline hydrolysis as described previously (Munn et al., 1999; Heese-Peck et al., 2002). Total sterol extracts were taken up in 10 μl pyridine and silylated using 10 μl of BSTFA (Sigma, St. Louis, MO). For gas-liquid chromatography–mass spectrometry (GC-MS) samples were diluted in ethyl acetate and analyzed on an HP 5890 Series II gas chromatograph (Hewlett-Packard, Palo Alto, CA) equipped with an HP 5972 mass selective detector and an HP 5-MS column (cross-linked 5% phenyl methyl siloxane; dimensions 30 m × 0.25 mm ×0.25-μm film thickness). Aliquots of 1 μl were injected in the splitless mode at 270°C injection temperature with helium as carrier gas at a flow rate of 0.9 ml/min in constant flow mode. The temperature program was 1 min at 100°C, 10°C/min to 250°C, and 3°C/min to 310°C. Mass spectra were acquired in scan mode (range, 200–550 atomic mass units) with 3.27 scans per second. Sterols were identified based on their mass fragmentation pattern and retention time relative to the cholesterol standard (Mullner et al., 2005).
Fluorescence Anisotropy Measurements
Cells were grown in YPD medium at 30°C to 1–5 × 107 cells/ml. After two washes (10 mM Tris-Cl, 1 mM EDTA, pH 7.0), they were labeled with 0.5 μM of 1-[4-(trimethylamino)pheny]-6-phenyl-1,3,5-hexatriene (TMA-DPH; Invitrogen, Carlsbad, CA) at 25°C for 10 min. The sur2 strains were labeled with 2 μM TMA-DPH. After two washes, the cells were placed in a RF-5300PC spectrofluorometer (Shimadzu, Kyoto, Japan) fixed with a polarization unit. The excitation and emission wavelengths were chosen at 358 and 458 nm and measured 25°C. Anisotropy (r) was calculated from polarized intensities according to the equation:
where I stands for fluorescence intensity of TMA-DPH–labeled cells after subtraction of that of unlabeled cells. The subscript VV indicates measurements with both polarizers positioned vertically, whereas subscript VH indicates measurements with the excitation polarizer in the vertical and the emission polarizer in the horizontal position. G stands for the G-factor, IHV/IHH, for this instrument, where subscripts HV and HH indicate positions of polarizers which are the opposite of VH and VV, respectively. Data are represented as means ± SDs from three independent experiments.
Fluorescent Microscopy Analysis
Yeast strains with plasmid pRS416-TAT2-mRFP (generated by subcloning the HindIII-XbaI fragment from pMO10 (Okamoto et al., 2006) into pRS416 were grown to log phase at 25°C in synthetic minimal medium lacking uracil and tryptophan. TAT2- monomeric red fluorescent protein (mRFP) was observed with a fluorescence microscope (BX51; Olympus, Tokyo, Japan). The fluorescence intensity of TAT2-mRFP in the plasma membrane and vacuole was quantified by a Slide Book 4 Digital Microscopy software (Intelligent Imaging Innovations, Denver, CO) and expressed as a percentage of total intensity. The data are shown as means ± SDs from n = 27–59 cells. Yeast strains with plasmid Can1-green fluorescent protein (GFP) were grown in minimum media at 30°C to exponential phase (OD600 = 0.7) and resuspended in potassium phosphate buffer (pH 7.0). Live specimens were visualized using Zeiss 510 Meta confocal microscope (Thornwood, NY) with a 100× PlanApochromat objective (excitation 488 nm). Image processing was done with Image J (http://rsb.info.nih.gov/ij/). To examine patches on the plasma membrane Z-stacks were created to generate images of entire cells and confocal sections of the cell surface were visualized (data not shown). Fluorescence images of Pdr12-CFP were all acquired using the same exposure time.
Transcript Analysis
Yeast strains were grown in triplicate in exactly the same conditions (unique batch of YPUAD, same day) until an OD600 of 0.8–1.0 was reached. Two ml of culture was centrifuged; cells were frozen in liquid N2 and stored at −80°C. Total RNA extraction was performed using the RNeasy Mini Kit (No. 74104; Qiagen, Chatsworth, CA) mechanical disruption protocol including a column DNAse treatment.
RNA purity was controlled on a spectrophotometer; the ratios A260/A280 (protein contamination) and A260/A230 (guanidinium thiocyanate contamination) were more than or equal to 2.0. RNA integrity control was performed by capillary electrophoresis on an Agilent Technologies 2100 Bioanalyzer (Wilmington, DE).
Five hundred nanograms of RNA was amplified and labeled using MessageAmp II-Biotin Enhanced (No. AM1791; Ambion, Austin, TX). Biotin-labeled cRNA, 20 μg, was fragmented and hybridized for 16 h to Affymetrix GeneChip Yeast2.0 Array (Affymetrix, Santa Clara, CA). The hybridization and development with streptavidin was performed using GeneChip Hybridization Wash and Stain Kit (No. 900720; Affymetrix). Washing and development of the chip was done on GeneChip Fluidics Station 450. Three biological replicates were processed for each strain.
Microarray Analysis
Summarization and normalization (scaling) of the microarray data were conducted using GCOS (Affymetrix). The software package GeneSpring GX 7.3 Analysis Platform (Agilent Technologies) and Matlab 2008 (MathWorks, Natick, MA) were then used to analyze the microarray data. Microarray expression ratios were presented as log-transformed base 2. Hierarchical clustering analysis focused on transcript expression patterns rather than absolute values and the triplicate data over each strain was averaged using the mean in the log2 transform matrix. The average of each strain (i.e., every probeset) was then subtracted from the averaged value of the wild-type strain in the gene expression matrix. The rows were then clustered using a correlation similarity metric and the average linkage method in Matlab.
RESULTS
Sterol Biosynthesis Mutants Show Altered Sphingolipid Profiles
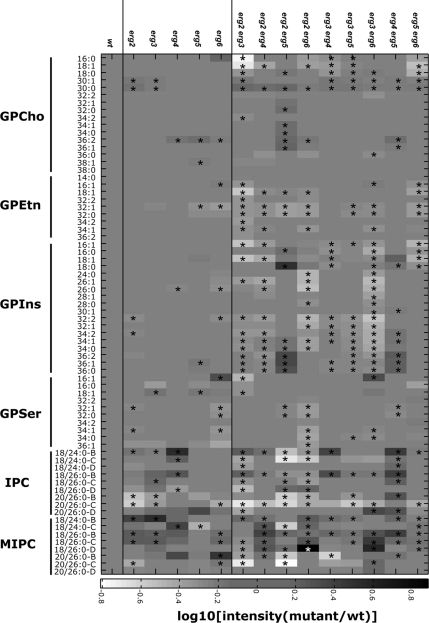
Mutants in the last five steps of ergosterol synthesis (Figure 1; Supplementary Figure S1) lead to a wide variety of phenotypes. We have created an isogenic set of single and double mutants in ergosterol biosynthesis and have characterized them for their sterol compositions and endocytic phenotypes (Munn et al., 1999; Heese-Peck et al., 2002). We noticed that the double erg3 erg6 mutant, which has a substantial growth defect, showed changes in its sphingolipid metabolism (Supplementary Figure S2) and that this change could be reversed by mutation of the yeast sphingomyelinase homolog (Sawai et al., 2000) encoded by ISC1 (Figure 1). The mutation in ISC1 aggravated the growth defect of the erg3 erg6 double mutant at 37°C (Supplementary Figure S2). These data suggest a functional interaction between sterols and sphingolipids, but this particular case might not be representative. Therefore, we systematically examined the lipid composition of all of our erg mutants using electrospray ionization–mass spectrometry (ESI-MS) techniques. The erg mutants (Supplementary Table S1) were grown in rich medium to log phase, harvested, washed, and frozen. Internal standards for quantitation were added, and the lipids were extracted and quantified by ESI-MS (Guan and Wenk, 2006). The quantification of the major glycerophospholipids (except for cardiolipin) and several of the major sphingolipids are presented. Results on mannosyldiinositolphosphorylceramide (M(IP)2C) are not presented because the data were not easily interpretable because of the presence of singly and doubly charged species, together with the fact that no appropriate internal standard was available. By steady-state phosphate or inositol labeling and thin-layer chromatography analysis, M(IP)2C represents ∼20% of the total sphingolipids in Saccharomyces cerevisiae (Schorling et al., 2001). Quantification was carried out relative to wild-type cells and is presented in Figure 2. The single erg mutants showed substantial changes (>30%) in <10% of their glycerophospholipid species, but showed substantial changes in more than 45% of their sphingolipid species. Remarkably, the sphingolipid pattern found in each single erg mutant was different. Therefore, the changes cannot be due to a lack of ergosterol because none of the mutant strains have ergosterol, but should be due to the presence of the abnormal sterols. This astounding result implies that yeast cells have the capacity to modify their sphingolipid composition in response to changes in sterol structures in their membranes. Many of the sterols accumulated in the mutants represent natural biosynthetic intermediates of ergosterol biosynthesis and are also found at low levels in wild-type cells (Supplementary Table S2). It is unlikely that cells are reacting to the concentration of sterols in the membrane because the total amount of sterols present in the single erg mutants was always higher than in the wild-type (Supplementary Table 2).
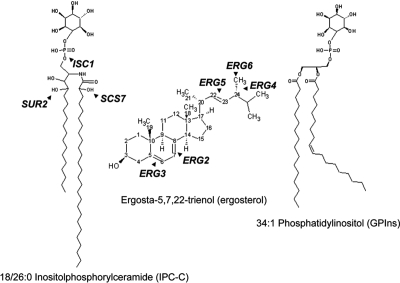
Figure 1.
Structures of some abundant sphingolipid, sterol, and glycerophospholipid species in S. cerevisiae. ISC1 encodes an inositolphosphorylceramide (IPC) hydrolase that converts IPC to ceramide and inositol phosphate. SUR2 and SCS7 encode enzymes responsible for hydroxylation of the sphingoid base and fatty acid, respectively, of sphingolipids. The last five steps of ergosterol biosynthesis are shown. The order of events is not implied here. For comparison, we show one of the most abundant yeast glycerophospholipids, 34:1 phosphatidylinositol. For details see Supplementary Figure S1. Structures were drawn with ACD/ChemSketch freeware.
Figure 2.
Lipidome of mutants in ergosterol biosynthesis. Isogenic wild-type and ergosterol mutant strains were grown as indicated. Lipid standards were added to 50 OD600-equivalents of cells, and lipids were extracted and measured using ESI-MS (Guan and Wenk, 2006). The quantities of lipids are expressed as ion intensities relative to wild-type levels, converted to a log10 scale. Glycerophospholipids: GPCho, glycerophosphocholine; GPEtn, glycerophosphoethanolamine; GPIns, glycerophosphoinositol; GPSer, glycerophosphoserine. Sphingolipids: IPC, inositol phosphorylceramide; MIPC, mannosyl inositol phosphorylceramide. The suffixes -B, -C, and -D on IPC and MIPC denote hydroxylation states, having two, three, or four hydroxyl groups, respectively. Data are presented as means of three independent biological samples. Statistical significance between wild-type and individual genotypes was determined using the Kruskal Wallis test. * p < 0.05.
Double mutants in ergosterol biosynthesis have substantial changes in their sphingolipid pattern, but also have substantially more changes in their glycerophospholipids (Figure 2). The single erg6 mutant appears somewhat intermediate in its lipid pattern between the single and double erg mutants. This is probably due to the fact that the ERG2 gene product works very inefficiently in the erg6 mutant strain, as noted by the accumulation of sterols with a double bond at position 8 (Heese-Peck et al., 2002; Supplementary Table S2E), explaining why the erg6 mutant has partial properties of a double erg2 erg6 mutant. Consistent with the radioactive labeling experiments, most of the double mutants as well as the erg2 and erg6 strains have less of the major inositolphosphoceramide (IPC) species, IPC-C. Otherwise, the patterns of sphingolipid prevalence vary considerably. Interestingly, when mutations in sphingolipid hydroxylation or head group turnover are introduced, there are no major changes in sterol compositions or quantities (Supplementary Table S2A). Most double erg mutants show more substantial changes in their sphingolipid patterns than either of the corresponding single erg mutants. There is one notable exception to this, the erg5 erg6 mutant, whose sphingolipid pattern is closer to wild type than either of the two single mutants. This might have evolutionary significance (see Discussion).
Sterol and Sphingolipid Biosynthesis Pathways Interact Genetically
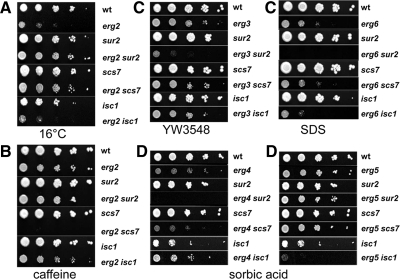
The specificity of the sphingolipid patterns in the different single erg mutants suggests that these changes may be a physiological response to accumulation of sterol biosynthetic intermediates and led us to hypothesize that sterols and sphingolipids functionally interact in yeast cells. To test this hypothesis, we created isogenic double mutants in sterol and sphingolipid biosynthesis. The prediction is that simultaneous defects in both pathways should lead to synthetic phenotypes. The changes in sphingolipid patterns (Figure 2) are mainly associated with changes in sphingolipid hydroxylation or in head group composition. Therefore, we chose a mutant that affects sphingolipid head group turnover, isc1, and mutants affecting sphingolipid hydroxylation. The sur2 mutant is unable to hydroxylate the sphingoid base and the scs7 mutant is defective in hydroxylation of the fatty acid on sphingolipids (see Figure 1 and Supplementary Figure S1; Haak et al., 1997). We made all 15 possible double mutant combinations of the five single erg mutants, with the three mutants affecting sphingolipid metabolism, and analyzed their ability to grow under a number of conditions. All of the single erg mutants, with or without sphingolipid defects, grew well at 30°C on rich medium, as do all of the double erg mutants except erg2 erg6 (Figure 3). At 37°C or on plates with nonfermentable carbon sources, several double mutants showed reduced growth when compared with the corresponding single mutants (marked with arrows). The double erg mutants notably exhibited more severe phenotypes than the single erg mutants from which they were derived.
Figure 3.
Systematic phenotypic analysis. The indicated strains were grown and pinned onto YPD or YPEG (1% yeast extract, 2% peptone, 3% ethanol, 3% glycerol, and 40 mM MES, pH 5.5) plates and grown at 30°C except when indicated. Plates were photographed after 2 (30 and 37°C) or 4 d (YPEG). Arrows denote conditions where sterol–sphingolipid double mutants showed different growth than the single erg mutants under the conditions shown. Each asterisk represents where the double mutant shows a change in growth properties (synthetic growth defect or suppression) when compared with the single erg mutant (Supplementary Figure S3). Of the 15 double mutants constructed, 13 showed synthetic phenotypes.
The mutants were next assayed for growth under a wide variety of conditions in order to uncover new phenotypes (Supplementary Figure S3). Some particularly illustrative examples from these assays are shown. Blocking hydroxylation of either the sphingoid base or the fatty acid of sphingolipids is able to suppress the slow growth phenotype of the erg2 mutant at 16°C (Figure 4A). The results found at 16°C are quite atypical, because changes in hydroxylation usually have much more specific effects on strains with aberrant sterols (Figure 4B). Combination of a mutation in sphingolipid fatty acid hydroxylation (scs7) with the erg2 mutation completely abrogates growth on plates with low amounts of caffeine, an inhibitor of the Target of Rapamycin Complex (TORC) 1 signaling pathway (Reinke et al., 2006), whereas both corresponding single mutants grow well on caffeine plates. A similar result was found for sensitivity to rapamycin (see below). Combination of sur2 with erg3 or erg6 led to synthetic growth phenotypes on plates with a low concentration of a glycosylphosphatidylinositol synthesis inhibitor, YW3548 (Sutterlin et al., 1997) or a low concentration of SDS (Figure 4C, left and right, respectively). These conditions are both expected to induce cell wall defects. The specificity of the site of hydroxylation in combination with particular sterol biosynthesis mutations suggest that it is the combination of the structures that leads to the synthetic phenotype. The simplest way to interpret this would be to postulate that sphingolipids and sterols interact physically in membranes to carry out some of their required functions. At the very least, we can conclude that they work in common pathways.
Figure 4.
Examples of suppression and synthetic phenotypes. Dilutions of indicated strains were spotted onto plates under the following conditions; 16°C (A), 2 mM caffeine (B), 1 μg/ml YW3548 (C, left), 0.01% SDS (C, right) or 1 mM sorbic acid at pH 4.5 (D).
Sorbic acid resistance is conferred by an ATP-dependent pump, Pdr12p (Piper et al., 1998), which exports weak acids such as sorbic and benzoic acids. This process is most affected by the erg3 and erg6 mutations (Supplementary Figure S3), suggesting that a Δ5 double bond in the B-ring and methylation of the side chain of ergosterol are the most important features for Pdr12p function. The erg4 mutant strain grows weakly on 1 mM sorbic acid plates, but combination with the sur2 mutation completely abrogates growth (Figure 4D). Introduction of the isc1 mutation improves growth slightly under the same conditions. The effects of sphingolipid changes on the erg5 mutant are quite different. The sur2 and scs7 mutations have almost no effect on growth of erg5 cells on sorbic acid plates, whereas introduction of the isc1 mutation completely blocks growth under the same conditions. These data show that the isc1 mutation is not having a simple positive or negative effect that is added to the effect of the sterol mutant. These results further confirm that it is the specific combinations of sphingolipid and sterol structures that lead to the synthetic phenotypes.
By using very simple tests we found synthetic phenotypes in 13 of the 15 possible double mutants in ergosterol and sphingolipid biosynthesis (Figure 3), providing genetic proof that these two pathways work together to carry out multiple cellular functions. A possible explanation for the genetic interaction could be that creation of the double mutants causes additional changes in glycerophospholipids and/or sterols. To examine this, we analyzed the glycerophospholipids, sphingolipid, and sterol compositions in the double mutant strains by ESI-MS. As expected from the introduction of mutations in sphingolipid modification, we observed dramatic changes in the sphingolipid pattern, but relatively minor changes in the phospholipids or sterols (Supplementary Figure S4 and Supplementary Table S2), lending additional support to our conclusion that it is the structural interactions between the sterols and sphingolipids in the membrane that cause the synthetic phenotypes and not further changes in phospholipids or sterols.
In this study, we have established that sterols and sphingolipids function together. We next sought to characterize the lipid dependence of two phenotypes to demonstrate how the combination of sterols and sphingolipids can affect the manner in which cells respond to drugs. The first example is the hypersensitivity of sterol–sphingolipid mutants to caffeine and rapamycin, and the second example is the hypersensitivity to sorbic acid.
Reduced TORC2 Activity Causes Caffeine and Rapamycin Hypersensitivity
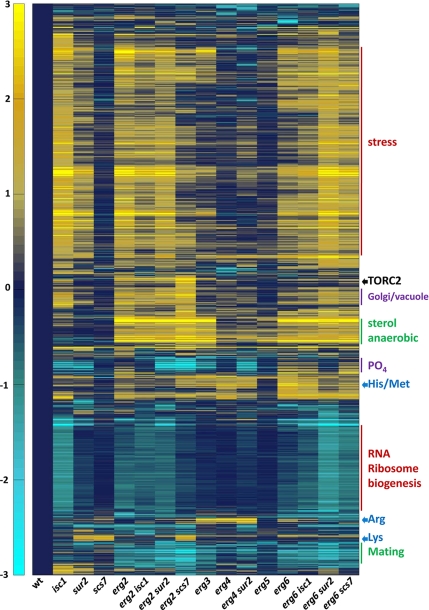
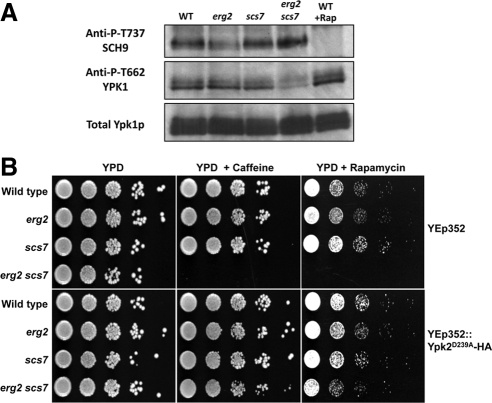
TORC1 is the target of caffeine and rapamycin in yeast both in vivo and in vitro (Wanke et al., 2008), whereas TORC2 is not significantly inhibited by either drug. Inhibition of TORC1 yields profound changes in the transcriptome, including a striking down-regulation of genes encoding ribosomal proteins (Martin et al., 2004; De Virgilio and Loewith, 2006). Therefore, we first examined expression of ribosomal protein mRNAs in our different strains to determine if reduced TORC1 activity could explain the increased caffeine sensitivity. Levels of mRNAs encoding ribosomal proteins were not affected in any of the mutants where transcript profiles were examined, including erg2 scs7 cells (see data at ArrayExpress at EBI, Experiment Riezman, Accession no. ID E-TABM-544). Surprisingly, when genes that were specifically overexpressed in the erg2 scs7 mutant were examined, we found RTA1, a gene that seems to be repressed by TORC2 (Mulet et al., 2006). On further inspection, a tight cluster of genes that are negatively regulated by TOR2 was found to be overexpressed in the erg2 scs7 mutant specifically (see Figure 7 and Supplementary Figure S5), suggesting that TORC2 might be less active in the mutant. This is the case because Ypk1p, one of the two Ypk kinases directly phosphorylated by TORC2 (Kamada et al., 2005; M. Picolis and R. Loewith, unpublished data) is hypophosphorylated in the mutant (Figure 5A). Sch9p, a direct TORC1 target (Urban et al., 2007), is phosphorylated normally in these strains, confirming that TORC1 is active. These results and the previous observation that ypk1 mutant cells are hypersensitive to rapamycin (Gelperin et al., 2002) suggest that reduced TORC2 activity and hypersensitivity to caffeine and rapamycin might be related. To investigate this, we introduced a plasmid carrying a constitutively active Ypk2 allele (Kamada et al., 2005) to bypass TORC2 function into the set of erg2 and scs7 mutant strains. The results were very clear. Ypk2 kinase activity suppressed the hypersensitivity of the erg2 scs7 strain to caffeine and rapamycin (Figure 5B). These results demonstrate that combination of the erg2 and scs7 mutations renders yeast cells hypersensitive to caffeine and rapamycin by lowering TORC2 activity. Strikingly, the sterol–sphingolipid composition affects how cells respond to these two drugs, not by affecting the direct target of the drug, but by affecting a partially overlapping pathway.
Figure 7.
Cluster map of transcripts that change in the sterol and sphingolipid mutants. Transcript levels were determined in the indicated strains. Data for transcripts that changed at least twofold under one condition were clustered. Predominant characteristics of gene clusters are indicated on the right. An expandable version of the figure with gene names or identifiers is provided (Supplementary Figure S6). The scale is a log transformed base 2.
Figure 5.
Hypersensitivity to caffeine and rapamycin is due to decrease in TORC2 activity. (A) Protein extracts were prepared from log phase cultures of wild-type, erg2, scs7, and erg2 scs7 mutant strains. The extent of phosphorylation of the hydrophobic motifs in Ypk1 and Sch9 (indicative of TORC2 and TORC1 activity, respectively) were determined by SDS-PAGE and Western blotting. Rapamycin treatment was for 20 min with 200 ng/ml. B) The same yeast strains, transformed with empty vector (Yep352) or an activated allele of Ypk2p (Yep352::Ypk2D239A-HA were grown to stationary phase, diluted, and 10-fold serial dilutions were plated onto plates containing 2 mM caffeine or 20 ng/ml rapamycin.
Reduced Activity of Pdr12p Causes Sorbic Acid Sensitivity
Sorbic acid is a commonly used food preservative that diffuses into cells when protonated at low pH and is trapped internally when it becomes deprotonated. It can be detoxified by yeast cells via the multidrug resistance (MDR)-like transporter, Pdr12p (Piper et al., 1998) in an ATP-dependent manner. To examine Pdr12p localization and function, we constructed a C-terminal genomic fusion creating a functional, fluorescently labeled Pdr12::CFP. Sorbic acid induces Pdr12p expression, and the protein is delivered to the cell surface (Piper et al., 1998) where it is the major, if not only protein that exports weak organic acids, including sorbic and benzoic acids as well as fluorescein (Holyoak et al., 1999). In wild-type, erg4, sur2, and erg4 sur2 mutants, Pdr12-CFP was induced and transported to the cell surface in approximately equal amounts as determined by fluorescence microscopy (Figure 6A). Pdr12p activity was assayed by measuring export of fluorescein. Fluorescein diacetate was loaded into yeast in the absence of glucose, conditions where export is defective. Glucose was added to allow fluorescein export. The double erg4 sur2 mutant showed drastically reduced fluorescein export, approximately six times less efficient than wild-type cells (Figure 6B). These results show that the MDR-like protein, Pdr12p, is properly transported to the cell surface, but is not active if membranes lack functional sterol–sphingolipid structures. In this example, sterol–sphingolipid composition seems to affect the drug export protein in a fairly direct manner.
Figure 6.
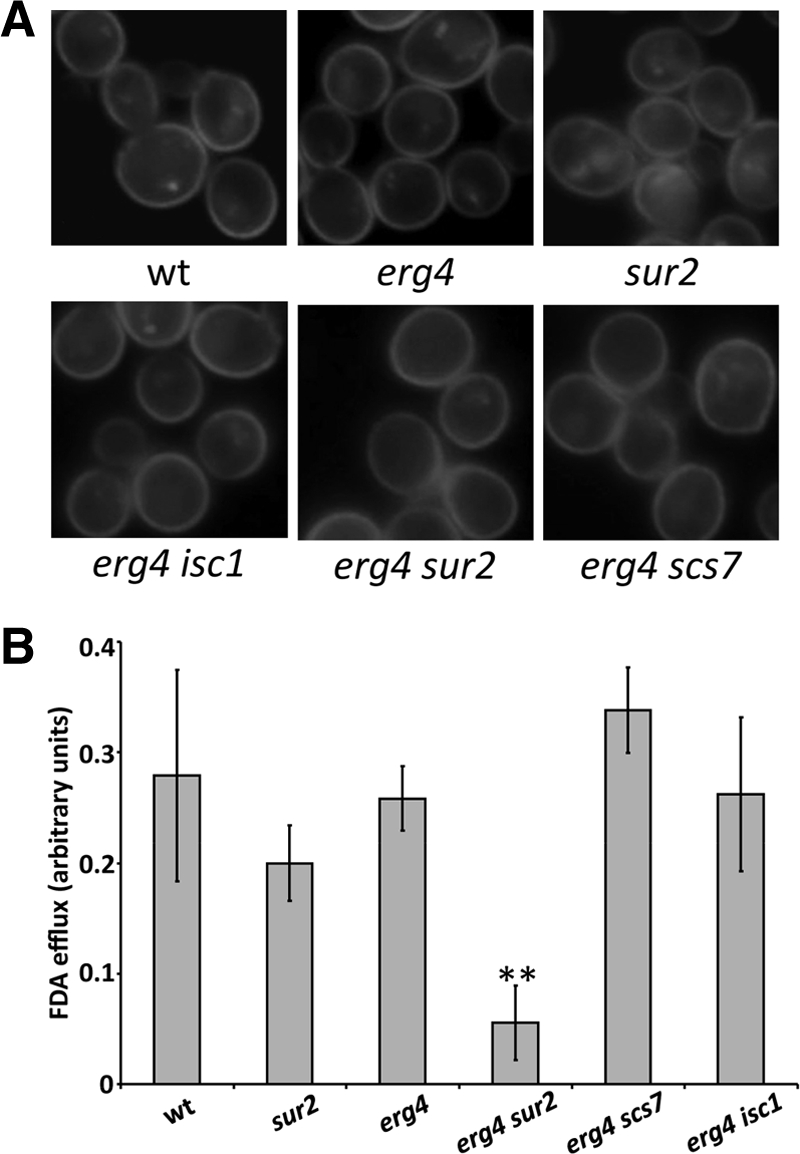
Sorbic acid sensitivity in erg4 sur2 is due to defective export by Pdr12p. (A) Pdr12p-CFP was visualized in the indicated strains. (B) The same strains were loaded with fluorescein diacetate in the presence of 2-deoxygluose to deplete ATP. The cells were harvested and resuspended in glucose containing medium and extracellular fluorescein was quantified after 15 and 30 s. Relative rates of FDA export per minute are shown. * p < 0.05 by the t test for mean.
Plasma Membrane Transporters and Anisotropy
Next, we turned our attention to protein trafficking where raft structures have been proposed to play an important role. We examined all of the single and double mutants and tested whether membrane protein-trafficking defects correlated with anisotropy measurements of the plasma membrane. One of the best established systems where proteins have been localized to specific plasma membrane microdomains is in S. cerevisiae, where two distinct types of domains have been seen. One of these is patches of ∼300 nm on the cell surface that contain the various permeases, including the tryptophan transporter Tat2p and the arginine transporter Can1p. Both transporters have been reported to be resistant to cold-detergent extraction, a technique that is often correlated with presence in rafts (Malinska et al., 2003, 2004; Umebayashi and Nakano, 2003; Mayor and Riezman, 2004). The permease-containing plasma membrane patches also stain for filipin, indicating the presence of ergosterol (Malinska et al., 2003, 2004). More recently, it has been shown that many genes, including some involved in sterol metabolism, affect the protein composition and structure of these patches (Grossmann et al., 2008). The permease-containing patches are surrounded by membrane domains containing the plasma membrane ATPase, a protein that has also been claimed to be functionally associated with rafts for biosynthetic delivery to the cell surface (Bagnat et al., 2000; Bagnat and Simons, 2002). Therefore, we analyzed the localization of fluorescently tagged Tat2p and Can1p, proteins found in the same domains at the cell surface. In wild-type cells under our conditions, Tat2p is found in approximately equal amounts in the plasma membrane and in the vacuole. The vacuolar Tat2p may be generated by direct targeting from the biosynthetic pathway or via delivery to the cell surface and subsequent endocytosis. Cell surface and vacuolar Tat2-GFP in the various mutants were quantified (Supplementary Figure S5; Table 1). The single erg mutants as well as isc1 and sur2 increased the amount of Tat2-GFP in the vacuole, and several strains showed slight endoplasmic reticulum (ER) accumulation. The increase in vacuole localization in the erg3 mutant is partially suppressed by the scs7 mutation, whereas the increase in vacuolar localization in the erg6 mutant is enhanced by isc1 and sur2 mutations. This demonstrates suppression and synthetic effects also for protein localization. On the other hand, the effects on Can1-GFP localization are only minor. In all mutants we detected the majority of Can1-GFP at the plasma membrane (Supplementary Figure S5). In the erg2 and erg3 series we found a small amount of vacuole staining, but no synthetic phenotypes (Table 1). The Can1-GFP construct was active in all strains because it conferred canavanine sensitivity. Therefore, two proteins localizing to the same plasma membrane patches seem to have quite different sterol and sphingolipid requirements.
Table 1.
Summary of Tat2p and Can1p localization data
| Genotype | Tat2 localization |
Can1p localization |
|||||
|---|---|---|---|---|---|---|---|
| PM | Vacuole | SD | PM | Vac | Small patches | Large patches | |
| wt | 46 | 54 | 6 | Mostly | No | Yes | Yes |
| isc1 | 28 | 72 | 5 | Mostly | No | Yes | No |
| sur2 | 35 | 65 | 6 | Mostly | No | Yes | No |
| scs7 | 41 | 59 | 5 | Mostly | No | Yes | No |
| erg2 | 35 | 65 | 6 | Mostly | No | Yes | Yes |
| erg2 isc1 | 36 | 64 | 8 | Mostly | Yes | Yes | No |
| erg2 sur2 | 34 | 66 | 6 | Mostly | Yes | Yes | No |
| erg2 scs7 | 34 | 66 | 6 | Mostly | Yes | Yes | Yes |
| erg3 | 28 | 72 | 5 | Mostly | Yes | Yes | Yes |
| erg3 isc1 | 24 | 76 | 5 | Mostly | Yes | Yes | Yes |
| erg3 sur2 | 26 | 74 | 7 | Mostly | Yes | nd | nd |
| erg3 scs7 | 37 | 63 | 7 | Mostly | Yes | nd | nd |
| erg4 | 34 | 66 | 6 | Mostly | No | Yes | Yes |
| erg4 isc1 | 47 | 53 | 4 | Mostly | No | Yes | Yes |
| erg4 sur2 | 35 | 65 | 7 | Mostly | No | Yes | No |
| erg4 scs7 | 38 | 62 | 6 | Mostly | No | Yes | No |
| erg5 | 33 | 67 | 5 | Mostly | No | No | Mainly |
| erg5 isc1 | 36 | 64 | 5 | Mostly | No | Yes | Yes |
| erg5 sur2 | 37 | 63 | 4 | Mostly | No | Yes | Yes |
| erg5 scs7 | 35 | 65 | 5 | Mostly | No | Yes | Yes |
| erg6 | 35 | 65 | 6 | Mostly | No | Yes | Yes |
| erg6 isc1 | 24 | 76 | 6 | Mostly | No | Yes | Yes |
| erg6 sur2 | 24 | 76 | 6 | Mostly | No | Yes | Yes |
| erg6 scs7 | 34 | 66 | 7 | Mostly | No | Yes | Reduced |
Very recently, Walther's group has proposed that TORC2 localizes to a third membrane domain (Berchtold and Walther, 2009). It will be of interest to determine if the localization of TORC2 to this domain is specifically altered in erg2 scs7 cells.
Because most of the plasma membrane surface is covered by proteins that have been proposed to localize to rafts, we decided to measure the fluorescence anisotropy of the plasma membrane using TMA-DPH. This fluorescent probe inserts into the yeast plasma membrane and measurement of its anisotropy correlates with membrane rigidity or lipid order (Sharma, 2006). The lower the anisotropy is, the more fluid is the membrane. Many of the erg mutants showed significantly and substantially lower anisotropy than wild-type cells, in particular, erg6 (Table 2). Interestingly, all of the double mutants with erg6 showed increased anisotropy, whereas they showed a greater defect in Tat2p surface localization and frequently exhibited more severe phenotypes in other assays performed in this study (Supplementary Figure S3). These results indicate that changes in sterol and sphingolipid composition definitely do affect membrane fluidity; however, there was a lack of correlation between the magnitude of these changes with any other phenotypic parameter that we have measured. From this systematic and unbiased approach to examine a role for membrane order on processes that depend upon the sterol and sphingolipid composition, we have to conclude that the fluidity/rigidity (order) of the membrane is not the major determinant in the phenotypes that we measured. This calls into question whether the lipid order predicted to be induced by sterol–sphingolipid interactions in the plasma membrane plays an important physiological role.
Table 2.
Anisotropy measurements using TMA-DPH
| Genotype | Mean | SD | Significance |
|---|---|---|---|
| WT | 0.283 | 0.004 | |
| isc1 | 0.278 | 0.005 | p < 0.2 |
| scs7 | 0.284 | 0.011 | |
| sur2 | 0.270 | 0.015 | p < 0.2 |
| erg2 | 0.274 | 0.008 | p < 0.2 |
| erg2 isc1 | 0.267 | 0.014 | p < 0.2 |
| erg2 sur2 | 0.276 | 0.005 | p < 0.2 |
| erg2 scs7 | 0.275 | 0.010 | |
| erg3 | 0.276 | 0.009 | |
| erg3 isc1 | 0.270 | 0.015 | |
| erg3 sur2 | 0.273 | 0.006 | p < 0.1 |
| erg3 scs7 | 0.291 | 0.010 | |
| erg4 | 0.280 | 0.006 | |
| erg4 isc1 | 0.268 | 0.006 | p < 0.05 |
| erg4 sur2 | 0.291 | 0.012 | |
| erg4 scs7 | 0.278 | 0.008 | |
| erg5 | 0.276 | 0.004 | p < 0.2 |
| erg5 isc1 | 0.285 | 0.005 | |
| erg5 sur2 | 0.279 | 0.019 | |
| erg5 scs7 | 0.277 | 0.005 | p < 0.2 |
| erg6 | 0.249 | 0.015 | p < 0.05 |
| erg6 isc1 | 0.265 | 0.008 | p < 0.05 |
| erg6 sur2 | 0.270 | 0.008 | p < 0.1 |
| erg6 scs7 | 0.268 | 0.009 | p < 0.1 |
Transcriptome of Sterol and Sphingolipid Biosynthesis Mutants
To begin to understand the basis for some of the phenotypes we observed in this study and the mechanism whereby cells change their sphingolipid composition in response to their sterol compositions, we performed transcript analysis of wild-type cells and 15 of the single and double mutants. Many sets of genes were unaffected by the mutations. These include genes required for the general secretion machinery, unfolded protein response, cell cycle machinery, large and small ribosome subunits, and many genes required for vacuole biogenesis (see data at ArrayExpress at EBI, Accession ID E-TABM-544). We eliminated all genes that did not show at least a twofold change under one condition and then clustered the genes together by expression pattern in the different strains using the correlation distance algorithm (Figure 7). Different patterns can be distinguished. For example, one set of genes is overexpressed in all erg2, erg3, and erg6 mutants and is relatively unaffected by mutations in isc1, sur2, or scs7. This set is characterized by a large number of genes related to ergosterol biosynthesis and genes expressed during anaerobic growth, suggesting that these genes are coregulated by sterol composition. Another set of genes, involved in mating, shows the same pattern of regulation, but is underexpressed. The largest set, mainly comprising genes induced by stress responses (e.g., heat-shock proteins, trehalose synthesis, glycerol synthesis, and autophagy) is characterized by overexpression in isc1, erg2, and erg6 strains, but with only mild, if any, overexpression in the erg3, erg4, or erg5 strains. In this set, synthetic and suppressive effects due to combination of sterol and sphingolipid mutations can be seen. For example, strong overexpression of many stress genes are seen in isc1, erg2, and erg6, but combination of the isc1 mutation with either erg2 or erg6 usually results in less overexpression. The effect of the sur2 mutation on overexpression is less than the isc1 mutation. However, combination of sur2 with either erg2 or erg6 results in stronger overexpression than the corresponding isc1 double mutants. It is clear from these results that most of the phenotypes we have identified with our plate assays do not correlate with induction of stress genes and therefore are probably not an indirect result of cellular stress. Again, another set of genes, involved in ribosome biogenesis and RNA processing, shows the same pattern, but with underexpression. Genes negatively regulated by TORC2 are overexpressed most strongly in the erg2 scs7 mutant. Several genes regulating products stored in the yeast vacuole (phosphate, basic amino acids) are affected. Genes involved in phosphate metabolism and storage are strongly down-regulated by sphingolipid mutations and display a particularly strong phenotypic enhancement when the erg2 and scs7 mutations are combined. The erg3 mutation causes some down-regulation, but the erg6 mutation causes a small up-regulation. Two other groups that form clusters are genes involved in basic amino acid biosynthesis, but interestingly, genes required for arginine biosynthesis are regulated differently from those for lysine biosynthesis. Several other smaller clusters can be seen and are noted (Supplementary Figure 6).
We examined if changes in transcript profiles in the erg mutants could explain the changes in sphingolipid composition. Of the more than 20 genes involved in sphingolipid biosynthesis and degradation (Supplementary Figure S1), we found four genes whose transcript levels changed significantly. Three of these are involved in sphingolipid degradation. YDC1 and YPC1, the two ceramide hydrolases, are coregulated with stress genes. YSR3, one of two sphingoid base phosphate phosphatases is coregulated with sterol biosynthesis genes. Finally, SUR1, encoding the catalytic subunit of one of the two mannosyltransferases making MIPC, is up-regulated in all strains tested, except the scs7, erg3, and erg5 mutants. These results strongly suggest that the changes in sphingolipid composition seen in the erg mutants are not the results of transcript changes.
DISCUSSION
One of the major new findings emanating from this study is that cells have a mechanism to sense and react to changes in their membrane sterol composition by modifying their membrane lipid compositions affecting mainly sphingolipids. The mechanism of the adaptation of sphingolipid levels is not understood yet, but is most likely not uniquely due to differences in transcript levels of the different genes involved in the pathway as most of the genes were unchanged. The mechanism used to detect the changes in sterol composition is currently under study and genetic approaches are possible. It is not the absence of ergosterol that is sensed, because each erg mutant shows a different pattern of sphingolipids. The pathway leading to changes in sphingolipid composition is most likely fundamentally different from previously described sterol-sensing mechanisms where different amounts of cholesterol are sensed by the SREBP pathway or other sterol sensors and then control gene expression (Goldstein et al., 2006), because sterol intermediates, rather than the total quantity of sterols, seem to affect sphingolipid composition in different ways and the changes are unlikely brought about directly through changes in gene expression. Recently, an SREBP-dependent pathway has been described in Schizosaccharomyces pombe, which has several features in common with what we have seen here, because there is a coordinate regulation of anaerobically expressed genes with ergosterol biosynthesis genes (Todd et al., 2006; Figure 7) and because sterol biosynthesis intermediates seem to be important (Hughes et al., 2007), but no specific effects on sphingolipids have been described. The pathway cannot be identical in S. cerevisiae because this yeast does not have an SREBP homolog, although they do have oxysterol-binding proteins that have been proposed to be involved in intracellular sterol transport (Raychaudhuri et al., 2006).
An interesting question is the physiological significance of the adaptation mechanism. First, it is likely that natural intermediates in the ergosterol biosynthesis pathway are being recognized. They are present in significant amounts under normal growth conditions and are found in larger amounts in the erg mutants (Supplementary Table S2). Sterol biosynthetic intermediates have been shown to be localized preferentially to the ER membrane (Zinser et al., 1993), and their amounts could be important in regulating adaptation mechanisms. Second, yeast may frequently encounter azole and other inhibitors of ergosterol biosynthesis in their natural environment and therefore require adaptation mechanisms to adjust their sphingolipid composition when azole compounds are present and inhibit sterol biosynthesis at various steps.
The control of sphingolipid species is unlikely to be solely the result of transcriptional changes because few differences in sphingolipid metabolic enzymes were observed. In particular, some changes in sphingolipid content are likely to be due to head group turnover via Isc1p (Supplementary Figure S2). The mode of activation of Isc1p in this case is unknown, but one possibility is regulation by localization because its localization has been shown to change as a result of changes in culture conditions in yeast (Vaena de Avalos et al., 2004).
In contrast to the effect of sterol mutations on sphingolipid composition, mutations affecting hydroxylation and head group turnover of sphingolipids do not seem to affect the sterol composition. Therefore, this process is not reciprocal. There could be different explanations for this situation. First, the species of sphingolipids that change in the sterol mutants are sphingolipid species that are normally present in substantial quantities, and only their amount changes. They cannot all be considered as intermediates, because each of these sphingolipid species might be functional. Known antifungals that target the sphingolipid biosynthesis pathway act at or before the step of inositolphosphorylceramide (IPC) synthesis (Horvath et al., 1994; Mandala and Harris, 2000; Sugimoto et al., 2004). In contrast, the major sterol species is ergosterol, and other species are probably just intermediates in its biosynthesis. Second, during evolution some metazoans either never had or lost the ability to synthesize sterols and are sterol auxotrophs (Vinci et al., 2008). Two particularly well-studied systems are Drosophila and Caenorhabditis elegans (Silberkang et al., 1983; Matyash et al., 2001). Experiments with Drosophila did not detect any major changes in phospholipid head groups or acyl group composition when grown with different amounts of cholesterol (Silberkang et al., 1983). Sphingolipids were not examined in this study. No information is published yet from either system about the lipid composition of these organisms when they are grown using different sterols. It is possible that these organisms have a mechanism similar to that described here for yeast that could be used to adjust membrane composition in response to which sterols they ingest.
The specific and major changes in sphingolipid species when sterol intermediates are present in the membrane show a dependence of sphingolipid metabolism on sterol composition, but also suggest that sterols and sphingolipids function together. The second major finding from our study stems from our genetic analysis of double mutants in sterol and sphingolipid metabolism and provides proof for their functional interaction. This study revealed a number of novel synthetic and suppression phenotypes between sterol and sphingolipid biosynthesis mutants. Two aspects of this systematic and unbiased analysis are particularly convincing. First is the penetration of phenotypes, with 13 of the 15 possible double mutants showing synthetic or suppression phenotypes. Second is the astounding specificity of the combinations required to affect each phenotype and the strict specificity for the site of hydroxylation on the sphingolipid molecule and its combination with a particular sterol mutation. Synthetic phenotypes can occur for different reasons: when there are two parallel pathways and one mutation is in each pathway or when two mutations affect the same pathway, quite often at the same step. Each mutation would affect the step partially, but the double mutant would have a much stronger phenotype. Classically, this can result when two proteins work together as a complex. In a similar manner it is possible to interpret our data by suggesting that sterols and sphingolipids might function in a variety of pathways as a sterol–sphingolipid complex. Sterols and sphingolipids are clearly capable of forming condensed complexes in artificial membranes (Radhakrishnan et al., 2000), and there is no reason to expect that they should not be able to do the same in biological membranes.
We tested many different phenotypes including growth in presence of cell wall disturbances, weak organic acid sensitivity, different carbon sources, temperatures, various classes of inhibitors affecting a number of different pathways as well as a systematic analysis of transcript levels. It is virtually impossible that sterols and sphingolipids happen to act in parallel pathways to carry out each of these functions where synthetic phenotypes have been uncovered. Therefore, we are obliged to conclude that sterols and sphingolipids function together to carry out a wide variety of cellular functions. The conclusion that sterols and sphingolipids function together is consistent with one of the tenets of the raft hypothesis. The other main tenet of the raft hypothesis is that the increased order produced by sterol–sphingolipid interactions is important for function. To begin to test this, we measured plasma membrane anisotropy as an indicator of membrane order. Even though there were substantial and significant differences in anisotropy, indicating a decrease in membrane order in several of the mutants, we were not able to correlate the measurements of membrane anisotropy with any of the phenotypes we assayed, and therefore we conclude that membrane order is not likely to have a great influence on the functions we examined. Therefore, our results do not provide support for this part of the raft hypothesis.
The raft hypothesis has been invoked to explain a large number of membrane trafficking events, which prompted us to examine the localization of two plasma membrane proteins that have been shown to be localized in microdomains of the plasma membrane. Here again, the raft hypothesis does not easily help to explain the results of steady-state localization of Tat2p and Can1p in the various single and double mutants. The two transporters colocalize to the same plasma membrane patches, but they show completely different dependence on sterols and sphingolipids for their localization. Localization of Tat2p, which was affected by sphingolipid and sterol mutations, did not correlate with anisotropy measurements. It could be that anisotropy measurements of the plasma membrane do not reflect fluidity of the compartment where Tat2p sorting occurs (Golgi or endosomal compartment) or that measuring the overall fluidity of the membrane does not indicate the fluidity in rafts, so we cannot disprove involvement of liquid order phases. On the other hand, there is no reason from our studies or others in the literature at this time to invoke a role for liquid order phases in vivo.
In what ways could sterol–sphingolipid interactions influence function with such a complex pattern of phenotypes? A trivial explanation would be that addition of sterol mutations with sphingolipid mutations cause an indirect effect by generally increasing the stress to the cells, which affects many functions. This is not the case for the hypersensitivities seen here to caffeine or sorbic acid, which do not correlate with stress gene induction. Indirect effects, although certainly occurring in some cases, are unlikely to be a common explanation for the synthetic effects we see because of the specificity of the position of hydroxylation of the sphingolipid and because different combinations of specific sterol and sphingolipid mutations affect only specific cellular pathways rather than having highly pleiotropic effects.
Biophysical (Ahmed et al., 1997; Radhakrishnan et al., 2001; Feigenson, 2007) and molecular dynamics (Aittoniemi et al., 2007) experiments have clearly demonstrated that sterols and sphingolipids can interact preferentially in artificial membranes. This means that these lipids exist in at least two forms in the membrane, free sterol and sphingolipid, as well as sterol–sphingolipid complexes. Our results suggest that this interaction will also be seen in vivo if suitable techniques become available. We postulate that most of the phenotypes seen here are mainly the result of defects in direct protein–lipid interactions and not changes in the fluidity properties of the membrane or membrane domains. Any integral or peripheral membrane protein could interact with either a sterol, a sphingolipid, or a sphingolipid-sterol complex, helping to explain the complexity of the phenotypic analysis. Membrane proteins might require the sterols and/sphingolipids for proper folding, assembly, targeting, regulation, or activity. Eukaryotic membrane proteins have had a long time to optimize these interactions during evolution because most cells have sterols and all have sphingolipids. Direct sterol–sphingolipid interactions could also help to explain why it is so difficult to express, purify, and crystallize eukaryotic membrane proteins in bacteria.
There are various ways to interpret the enhancement of phenotypes we have seen. For example, in erg4 and sur2 single mutant cells Pdr12p shows substantial activity, but in erg4 sur2 double mutants activity is lost. Pdr12p might require interaction with sterol–sphingolipid complexes for full activity. The interaction could be reduced because of a stronger change in shape of the complex due to the combination. The complex might adapt a different tilt in the membrane because of the two mutations. Molecular dynamics studies suggest that sphingomyelin binding to cholesterol controls its tilt in the lipid bilayer (Aittoniemi et al., 2006). Alternatively, the combination of the erg4 and sur2 mutations could affect the equilibrium between the free lipids with sterol–sphingolipid complexes affecting Pdr12p activity. We propose that this paradigm—of protein interaction with sterols, sphingolipids, or their complexes—could explain the majority of phenotypes we see and that have been shown to depend upon sterols or sphingolipids in the literature without invoking the need for order in the membranes.
In order for sterols and sphingolipids to function together, their structural compatibility had to be maintained through evolution. Strikingly, there was one outlier from the lipid analysis of the double erg mutants (Figure 2). The sphingolipid pattern in the erg5 erg6 double mutant was more similar to wild type than the corresponding single mutants. The double mutation is “sphingolipid neutral.” The ERG5 and ERG6 genes are responsible for changes in the sterol side chain, the principle difference in the biosynthetic pathways of cholesterol and ergosterol. The two genes are linked in S. cerevisiae. This suggests that the ERG5 and ERG6 genes could have been inherited or lost together during evolution without causing major sphingolipid changes, allowing a coevolution of sphingolipids and sterols.
This study also demonstrates that changes in sterol and sphingolipid composition can affect how a cell responds to a drug or inhibitor. Several examples are shown here, most prominently, the hypersensitivity of the erg2 scs7 double mutant to caffeine and rapamycin. Our data show that the reduction in activity of TORC2, not TORC1, the direct target of caffeine and rapamycin, is the reason for the hypersensitivity. The extent to which this paradigm can be extended to humans is not presently clear, but it is evident that genetic factors and diet affect sensitivity and effects of certain drugs. Sterols and sphingolipid composition could play an important role in pharmacological variations in the population.
Supplementary Material
ACKNOWLEDGMENTS
We thank Erich Leitner for operating GC-MS measurements; Isabelle Riezman and Brigitte Bernadets for excellent technical assistance; Guanghou Shui, Gek Huey Chua, Reika Watanabe, Jean Gruenberg, and Marcos González-Gaitán for helpful discussions; Patrick Descombes and the Genomics Platform of the NCCR Genetics for help with the transcript profiling; Christoph Bauer and the Imaging Platform of the NCCR Genetics for help with microscopy; and Yoshifumi Jigami (National Institute of Advanced Industrial Science and Technology) for the gift of pMO10 plasmid. M.R.W. was supported in part by the Singapore National Research Foundation under CRP Award No. 2007-04, the Biomedical Research Council of Singapore (R-183-000-211-305) and the National Medical Research Council (R-183-000-224-213). H.R. was supported by grants from the Swiss National Science Foundation and the Human Frontiers Science Program Organization and a FEBS long-term fellowship (H.P.). This project was financed with a grant from the Swiss SystemsX.ch initiative, evaluated by the Swiss National Science Foundation.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-11-1126) on February 18, 2009.
REFERENCES
- Ahmed S. N., Brown D. A., London E. On the origin of sphingolipid/cholesterol-rich detergent-insoluble cell membranes: physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble, liquid-ordered lipid phase in model membranes. Biochemistry. 1997;36:10944–10953. doi: 10.1021/bi971167g. [DOI] [PubMed] [Google Scholar]
- Aittoniemi J., Niemela P. S., Hyvonen M. T., Karttunen M., Vattulainen I. Insight into the putative specific interactions between cholesterol, sphingomyelin, and palmitoyl-oleoyl phosphatidylcholine. Biophys. J. 2007;92:1125–1137. doi: 10.1529/biophysj.106.088427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aittoniemi J., Rog T., Niemela P., Pasenkiewicz-Gierula M., Karttunen M., Vattulainen I. Tilt: major factor in sterols' ordering capability in membranes. J. Phys. Chem. B Condens. Matter Mater. Surf. Interfaces Biophys. 2006;110:25562–25564. doi: 10.1021/jp064931u. [DOI] [PubMed] [Google Scholar]
- Anderson R. G., Jacobson K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 2002;296:1821–1825. doi: 10.1126/science.1068886. [DOI] [PubMed] [Google Scholar]
- Bagnat M., Keranen S., Shevchenko A., Shevchenko A., Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M., Simons K. Lipid rafts in protein sorting and cell polarity in budding yeast Saccharomyces cerevisiae. Biol. Chem. 2002;383:1475–1480. doi: 10.1515/BC.2002.169. [DOI] [PubMed] [Google Scholar]
- Baudry K., et al. The effect of the erg26–1 mutation on the regulation of lipid metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:12702–12711. doi: 10.1074/jbc.M100274200. [DOI] [PubMed] [Google Scholar]
- Berchtold D., Walther T. C. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol. Biol. Cell. 2009 doi: 10.1091/mbc.E08-10-1001. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher M. S., Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- De Virgilio C., Loewith R. Cell growth control: little eukaryotes make big contributions. Oncogene. 2006;25:6392–6415. doi: 10.1038/sj.onc.1209884. [DOI] [PubMed] [Google Scholar]
- Eisenkolb M., Zenzmaier C., Leitner E., Schneiter R. A specific structural requirement for ergosterol in long-chain fatty acid synthesis mutants important for maintaining raft domains in yeast. Mol. Biol. Cell. 2002;13:4414–4428. doi: 10.1091/mbc.E02-02-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigenson G. W. Phase boundaries and biological membranes. Annu. Rev. Biophys. Biomol. Struct. 2007;36:63–77. doi: 10.1146/annurev.biophys.36.040306.132721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin D., Horton L., DeChant A., Hensold J., Lemmon S. K. Loss of ypk1 function causes rapamycin sensitivity, inhibition of translation initiation and synthetic lethality in 14-3-3-deficient yeast. Genetics. 2002;161:1453–1464. doi: 10.1093/genetics/161.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J. L., DeBose-Boyd R. A., Brown M. S. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Grimm M. O., et al. Regulation of cholesterol and sphingomyelin metabolism by amyloid-beta and presenilin. Nat. Cell Biol. 2005;7:1118–1123. doi: 10.1038/ncb1313. [DOI] [PubMed] [Google Scholar]
- Grossmann G., Malinsky J., Stahlschmidt W., Loibl M., Weig-Meckl I., Frommer W. B., Opekarova M., Tanner W. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 2008;183:1075–1088. doi: 10.1083/jcb.200806035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X. L., Wenk M. R. Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from Saccharomyces cerevisiae. Yeast. 2006;23:465–477. doi: 10.1002/yea.1362. [DOI] [PubMed] [Google Scholar]
- Haak D., Gable K., Beeler T., Dunn T. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 1997;272:29704–29710. doi: 10.1074/jbc.272.47.29704. [DOI] [PubMed] [Google Scholar]
- Heese-Peck A., Pichler H., Zanolari B., Watanabe R., Daum G., Riezman H. Multiple functions of sterols in yeast endocytosis. Mol. Biol. Cell. 2002;13:2664–2680. doi: 10.1091/mbc.E02-04-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holyoak C. D., Bracey D., Piper P. W., Kuchler K., Coote P. J. The Saccharomyces cerevisiae weak-acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy-dependent mechanism. J. Bacteriol. 1999;181:4644–4652. doi: 10.1128/jb.181.15.4644-4652.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A., Sutterlin C., Manning-Krieg U., Movva N. R., Riezman H. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. L., Lee C. Y., Bien C. M., Espenshade P. J. 4-Methyl sterols regulate fission yeast SREBP-Scap under low oxygen and cell stress. J. Biol. Chem. 2007;282:24388–24396. doi: 10.1074/jbc.M701326200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K., Mouritsen O. G., Anderson R. G. Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- Jin H., McCaffery J. M., Grote E. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast. J. Cell Biol. 2008;180:813–826. doi: 10.1083/jcb.200705076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Fujioka Y., Suzuki N. N., Inagaki F., Wullschleger S., Loewith R., Hall M. N., Ohsumi Y. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 2005;25:7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., Kasai R. S., Kondo J., Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- Malinska K., Malinsky J., Opekarova M., Tanner W. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell. 2003;14:4427–4436. doi: 10.1091/mbc.E03-04-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinska K., Malinsky J., Opekarova M., Tanner W. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 2004;117:6031–6041. doi: 10.1242/jcs.01493. [DOI] [PubMed] [Google Scholar]
- Mandala S. M., Harris G. H. Isolation and characterization of novel inhibitors of sphingolipid synthesis: australifungin, viridiofungins, rustmicin, and khafrefungin. Methods Enzymol. 2000;311:335–348. doi: 10.1016/s0076-6879(00)11094-8. [DOI] [PubMed] [Google Scholar]
- Martin D. E., Soulard A., Hall M. N. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Matyash V., Entchev E. V., Mende F., Wilsch-Brauninger M., Thiele C., Schmidt A. W., Knolker H. J., Ward S., Kurzchalia T. V. Sterol-derived hormone(s) controls entry into diapause in Caenorhabditis elegans by consecutive activation of DAF-12 and DAF-16. PLoS Biol. 2004;2:e280. doi: 10.1371/journal.pbio.0020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V., Geier C., Henske A., Mukherjee S., Hirsh D., Thiele C., Grant B., Maxfield F. R., Kurzchalia T. V. Distribution and transport of cholesterol in Caenorhabditis elegans. Mol. Biol. Cell. 2001;12:1725–1736. doi: 10.1091/mbc.12.6.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S., Riezman H. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- Mitra K., Ubarretxena-Belandia I., Taguchi T., Warren G., Engelman D. M. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl. Acad. Sci. USA. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulet J. M., Martin D. E., Loewith R., Hall M. N. Mutual antagonism of target of rapamycin and calcineurin signaling. J. Biol. Chem. 2006;281:33000–33007. doi: 10.1074/jbc.M604244200. [DOI] [PubMed] [Google Scholar]
- Mullner H., Deutsch G., Leitner E., Ingolic E., Daum G. YEH2/YLR020c encodes a novel steryl ester hydrolase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:13321–13328. doi: 10.1074/jbc.M409914200. [DOI] [PubMed] [Google Scholar]
- Munn A. L., Heese-Peck A., Stevenson B. J., Pichler H., Riezman H. Specific sterols required for the internalization step of endocytosis in yeast. Mol. Biol. Cell. 1999;10:3943–3957. doi: 10.1091/mbc.10.11.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Yoko-o T., Umemura M., Jigami Y. Glycosylphosphatidylinositol-anchored proteins are required for the transport of detergent-resistant microdomain-associated membrane proteins Tat2p and Fur4p. J. Biol. Chem. 2006;281:4013–4023. doi: 10.1074/jbc.M504684200. [DOI] [PubMed] [Google Scholar]
- Pagano R. E., Puri V., Dominguez M., Marks D. L. Membrane traffic in sphingolipid storage diseases. Traffic. 2000;1:807–815. doi: 10.1034/j.1600-0854.2000.011101.x. [DOI] [PubMed] [Google Scholar]
- Piper P., Mahe Y., Thompson S., Pandjaitan R., Holyoak C., Egner R., Muhlbauer M., Coote P., Kuchler K. The pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 1998;17:4257–4265. doi: 10.1093/emboj/17.15.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri V., Watanabe R., Dominguez M., Sun X., Wheatley C. L., Marks D. L., Pagano R. E. Cholesterol modulates membrane traffic along the endocytic pathway in sphingolipid-storage diseases. Nat. Cell Biol. 1999;1:386–388. doi: 10.1038/14084. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A., Anderson T. G., McConnell H. M. Condensed complexes, rafts, and the chemical activity of cholesterol in membranes. Proc. Natl. Acad. Sci. USA. 2000;97:12422–12427. doi: 10.1073/pnas.220418097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A., Li X. M., Brown R. E., McConnell H. M. Stoichiometry of cholesterol-sphingomyelin condensed complexes in monolayers. Biochim. Biophys. Acta. 2001;1511:1–6. doi: 10.1016/s0005-2736(01)00274-7. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A., McConnell H. Condensed complexes in vesicles containing cholesterol and phospholipids. Proc. Natl. Acad. Sci. USA. 2005;102:12662–12666. doi: 10.1073/pnas.0506043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Im Y. J., Hurley J. H., Prinz W. A. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J. Cell Biol. 2006;173:107–119. doi: 10.1083/jcb.200510084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A., Chen J. C., Aronova S., Powers T. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J. Biol. Chem. 2006;281:31616–31626. doi: 10.1074/jbc.M603107200. [DOI] [PubMed] [Google Scholar]
- Rizzo M. A., Springer G. H., Granada B., Piston D. W. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Sawai H., Okamoto Y., Luberto C., Mao C., Bielawska A., Domae N., Hannun Y. A. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:39793–39798. doi: 10.1074/jbc.M007721200. [DOI] [PubMed] [Google Scholar]
- Scheek S., Brown M. S., Goldstein J. L. Sphingomyelin depletion in cultured cells blocks proteolysis of sterol regulatory element binding proteins at site 1. Proc. Natl. Acad. Sci. USA. 1997;94:11179–11183. doi: 10.1073/pnas.94.21.11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorling S., Vallee B., Barz W. P., Riezman H., Oesterhelt D. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. C. Implications of sterol structure for membrane lipid composition, fluidity and phospholipid asymmetry in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:1047–1051. doi: 10.1111/j.1567-1364.2006.00149.x. [DOI] [PubMed] [Google Scholar]
- Silberkang M., Havel C. M., Friend D. S., McCarthy B. J., Watson J. A. Isoprene synthesis in isolated embryonic Drosophila cells. I. Sterol-deficient eukaryotic cells. J. Biol. Chem. 1983;258:8503–8511. [PubMed] [Google Scholar]
- Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y., Sakoh H., Yamada K. IPC synthase as a useful target for antifungal drugs. Curr. Drug Targets Infect. Disord. 2004;4:311–322. doi: 10.2174/1568005043340597. [DOI] [PubMed] [Google Scholar]
- Sutterlin C., Horvath A., Gerold P., Schwarz R. T., Wang Y., Dreyfuss M., Riezman H. Identification of a species-specific inhibitor of glycosylphosphatidylinositol synthesis. EMBO J. 1997;16:6374–6383. doi: 10.1093/emboj/16.21.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd B. L., Stewart E. V., Burg J. S., Hughes A. L., Espenshade P. J. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol. Cell. Biol. 2006;26:2817–2831. doi: 10.1128/MCB.26.7.2817-2831.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umebayashi K., Nakano A. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 2003;161:1117–1131. doi: 10.1083/jcb.200303088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J., et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Vaena de Avalos S., Okamoto Y., Hannun Y. A. Activation and localization of inositol phosphosphingolipid phospholipase C, Isc1p, to the mitochondria during growth of Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:11537–11545. doi: 10.1074/jbc.M309586200. [DOI] [PubMed] [Google Scholar]
- Vance J. E. Lipid imbalance in the neurological disorder, Niemann-Pick C disease. FEBS Lett. 2006;580:5518–5524. doi: 10.1016/j.febslet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Vinci G., Xia X., Veitia R. A. Preservation of genes involved in sterol metabolism in cholesterol auxotrophs: facts and hypotheses. PLoS ONE. 2008;3:e2883. doi: 10.1371/journal.pone.0002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke V., Cameroni E., Uotila A., Piccolis M., Urban J., Loewith R., De Virgilio C. Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 2008;69:277–285. doi: 10.1111/j.1365-2958.2008.06292.x. [DOI] [PubMed] [Google Scholar]
- Wattenberg B. W., Silbert D. F. Sterol partitioning among intracellular membranes. Testing a model for cellular sterol distribution. J. Biol. Chem. 1983;258:2284–2289. [PubMed] [Google Scholar]
- Zinser E., Paltauf F., Daum G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J. Bacteriol. 1993;175:2853–2858. doi: 10.1128/jb.175.10.2853-2858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.