Abstract
Respiratory infection with highly pathogenic influenza A viruses is characterized by the exuberant production of cytokines and chemokines and the enhanced recruitment of innate inflammatory cells. Here, we show that challenging mice with virulent influenza A viruses, including currently circulating H5N1 strains, causes the increased selective accumulation of a particular dendritic cell subset, the tipDCs, in the pneumonic airways. These tipDCs are required for the further proliferation of influenza-specific CD8+ T cells in the infected lung, because blocking their recruitment in CCR2−/− mice decreases the numbers of CD8+ effectors and ultimately compromises virus clearance. However, diminution rather than total elimination of tipDC trafficking by treatment with the peroxisome proliferator-activated receptor-γ agonist pioglitazone moderates the potentially lethal consequences of excessive tipDC recruitment without abrogating CD8+ T cell expansion or compromising virus control. Targeting the tipDCs in this way thus offers possibilities for therapeutic intervention in the face of a catastrophic pandemic.
Keywords: H5N1, inflammation, pathogenesis
Influenza epidemics and pandemics vary greatly in pathogenicity. In any given year, the familiar “seasonal” influenza epidemic can be associated with >35,000 deaths in the United States alone (1), whereas the 1918 “Spanish flu” resulted in the deaths of ≥40 million people worldwide (2–7). With a global death rate exceeding 2.5%, the 1918 Spanish flu was the worst acute pandemic of modern times (8). Our current concern is with the avian H5N1 influenza A viruses. These virulent pathogens were first found to have crossed over from their avian reservoir in 1996 and as of January 2009 have been responsible for a 63% mortality rate in 399 laboratory-confirmed human cases (9). Unlike the situation for seasonal influenza where the immunocompromised (very young or elderly individuals) are most at risk, infection with the highly pathogenic (HP) 1918 H1N1 pandemic strain and recent H5N1 isolates is associated with high death rates in otherwise healthy, fully immunocompetent adults (8). This paradox led to the hypothesis that these HP influenza viruses induce immune-mediated pathology, characterized by severe vascular leakage and lung edema.
The idea that fatal respiratory compromise results from a dysregulated immune response, commonly referred to as a cytokine storm or hypercytokinemia, is supported by an expanding body of in vitro and in vivo evidence showing greatly elevated levels of proinflammatory mediators in the pneumonic lung (3, 5, 10–14). Here, we show that a subset of dendritic cells (DCs), described as TNF-α/inducible nitric oxide synthase (iNOS)-producing DCs (tipDCs) (15), accumulate in significantly greater numbers during the course of lethal (versus sublethal) influenza infections (Fig. S1). However, although it might be expected that eliminating the tipDCs would ameliorate the disease process, we found that the opposite was the case. The tipDCs also drive a local, protective CD8+“killer” T cell response in the virus-infected respiratory tract. Most interestingly, we established that partially compromising tipDC recruitment can be protective. Giving mice the type II diabetes drug pioglitazone diminishes but does not prevent tipDC recruitment, while allowing for sufficient CD8+ T cell expansion to protect against an otherwise lethal HP influenza virus challenge.
Results
Innate Cell Recruitment During HP and Sublethal Influenza Infections.
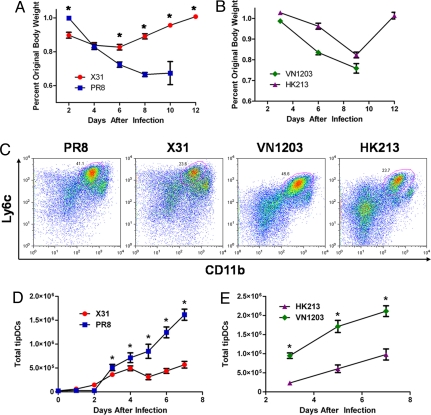
Although the expeditious accumulation of proinflammatory cytokines and chemokines during HP influenza infections has been extensively characterized, the identities of the cell types responsible for this immune-induced pathology are less clear. We thus measured lung recruitment kinetics for natural killer cells (NKs), conventional DCs (cDCs), neutrophils, macrophages, and tipDCs in C57BL/6J (B6) mice infected with either H1N1 A/Puerto Rico/8/34 (PR8), a mouse-adapted virus, or H3N2 A/Aichi/68 (x-31), a reassortant that contains the 6 internal genes of PR8 and the surface HA and neuraminidase (NA) from a prototypical H3N2 virus. Intranasal (i.n.) infection with 103 plaque-forming units (pfu) of PR8 is uniformly lethal by day 10 after infection, whereas all those given 105 pfu of x-31 recover by day 12 after losing 15–20% of their body weights (Fig. 1A). Tissues were collected daily (n = 5) for 7 days after i.n. challenge, and the inflammatory cell phenotypes were determined by flow cytometry (Table S1). The trafficking profiles for macrophages, cDCs, neutrophils, and NKs were comparable for the two viruses (Fig. S2). In contrast, the tipDCs accumulated to significantly higher numbers beginning on day 3 and increased through day 7 after infection with the HP PR8 virus (Fig. 1 C and D). Furthermore, as found by others (3, 5, 10–14), proinflammatory cytokine and chemokine levels were relatively elevated in the bronchoalveolar lavage (BAL) supernatant from mice given the HP virus (Fig. S3).
Fig. 1.
Increased pathology correlates with greater tipDC recruitment. (A and B) Host morbidity presented as mean of percentage original body weight ± SEM after infection with PR8 (lethal H1N1) or x-31 (sublethal H3N2) (A) and VN1203 (lethal H5N1) or HK213 (sublethal H5N1) (B). In general, mice infected with x-31 lose less weight than mice infected with PR8 (asterisks, simple effects ANOVA, P < 0.05). For the H5N1 viruses, HK213-infected mice lose less weight than VN1203-infected mice (virus effect, P < 0.0001). Results are representative of at least 3 independent experiments (n ≥ 5). (C) Representative FACS plots for total lung homogenate stained with mAbs to Ly6c and CD11b on day 5 after infection with PR8, x-31, VN1203, or HK213. The gated population (circled in red) from each plot represents tipDCs. (D and E) Mean number of tipDCs ± SEM recovered from total lung homogenate over time after infection with PR8 or x-31 (D) and VN1203 or HK213 (E). Sublethal infections resulted in fewer tipDCs in the lungs on days marked with an asterisk (simple effects ANOVA, P < 0.03). Results are representative of at least 3 independent experiments (n ≥ 5).
This analysis (Fig. 1 A–D) with x-31 and PR8 was then extended for two influenza viruses: H5N1 A/Vietnam/1203/2004 (VN1203, from a fatal human infection in 2004) has a LD50 of 1 pfu in B6 mice, whereas H5N1 A/Hong Kong/213/2003 (HK213) is much less virulent (LD50 = 103.7 pfu). Mice given 102 pfu of VN1203 were fatally compromised by day 9, whereas HK213 caused a 20% weight loss with full recovery by day 12 (Fig. 1B). As we discovered previously (Fig. 1 A–D), the tipDCs were found at significantly higher cell counts in the lungs of the HP VN1203-infected animals (Fig. 1 C and E), although there were no consistent differences in the numbers of macrophages, neutrophils, cDCs, or NKs (Fig. S4). Because of restrictions on the removal of samples from the high-security biosafety level (BSL) 3+ space where mice infected with these HP H5N1 viruses are held, we were not able to repeat the cytokine analyses shown for the x-31 and PR8 viruses in Fig. S3.
Chemokine (C-C Motif) Receptor 2 (CCR2) Regulates tipDC Trafficking and CD8+ T Cell Response Magnitude.
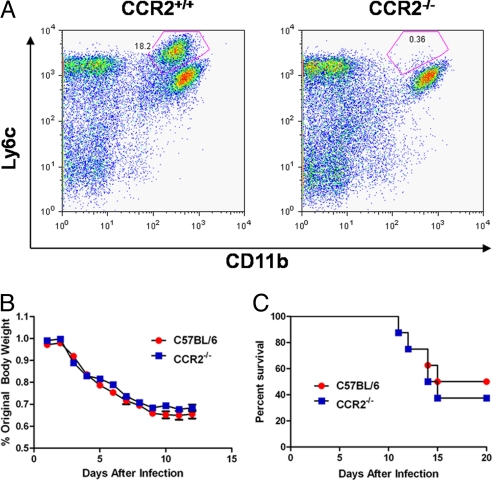
Emigration of tipDC precursors from the bone marrow is known to depend on signaling via the CCR2 receptor (16). We thus compared the trafficking kinetics of tipDCs between CCR2−/− and CCR2+/+ (B6) mice after infection with a sublethal dose (102 pfu) of PR8 virus. As described by others (16), few, if any, tipDCs could be recovered from the lungs of the CCR2−/− group (Fig. 2A). However, what was surprising in light of our previous conclusion that greater tipDC numbers in the airways correlate with a more severe disease profile (Fig. 1 C–E) was that the CCR2−/− mice showed no decrease in morbidity or mortality (Fig. 2 B and C) after HP virus challenge. These results are in contrast to a previous finding that CCR2 deficiency decreases influenza-induced mortality (17, 18). A comparison between the studies is difficult because each used a different model system. Dawson et al. (17) used an infectious dose of 5 hemagglutinating units that obscures the ability to compare infectious doses between studies. Lin et al. (18) used a tissue culture 50% infectious dose of 8.9 × 105. This dose is almost three logs higher than that used in the present study. It is reasonable that the noted differences in morbidity and mortality are due to the various infectious doses.
Fig. 2.
tipDCs fail to accumulate in the lungs of CCR2−/− animals. (A) Representative FACS plots of total lung homogenate from either CCR2+/+ (B6) or congenic CCR2−/− animals stained with mAbs to Ly6c and CD11b on day 5 after infection with 102 pfu of PR8 (sublethal infection). (B) Host morbidity presented as mean of percentage original body weight ± SEM after infection with 103 pfu of PR8 (lethal infection). Regression analysis did not reveal a significant difference in weight loss between the two strains of mice (strain effect, P = 0.22; interaction effect, P = 0.07). Results are representative of two independent experiments (n ≥ 5). (C) Elimination of tipDC recruitment to the lungs does not protect against lethal influenza challenge. Results of the Mantel–Cox test showed no statistically significant difference between groups (P = 0.66). Results are representative of 3 independent experiments (n ≥ 5).
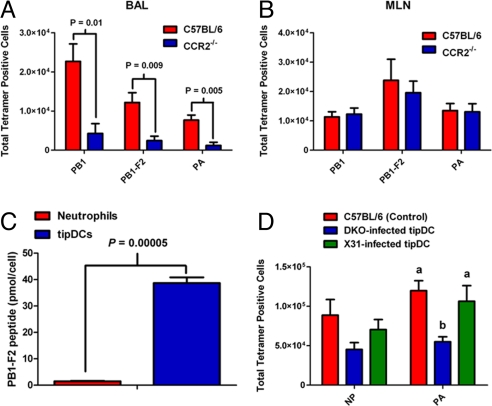
Further analysis suggested that the continued vulnerability in the absence of tipDCs might reflect a failure to control the infection in the lung. Although no virus could be recovered from the BAL fluid of the CCR2+/+ controls on day 9, a titer of 103.6 pfu (SEM ±103.3) was recorded in comparable samples from the CCR2−/− group. How might the presence or absence of tipDCs influence virus clearance? The primary mechanism for eliminating influenza-virus-producing respiratory epithelial cells is thought to be CD8+ cytotoxic T lymphocyte (CTL)-mediated lysis (19, 20). In addition to the effect on tipDC recruitment, there was (by staining with the DbPA224–233, KbPB1703–711, and DbPB1-F262–70 tetramers) a substantial reduction in influenza-specific CD8+ CTL counts for BAL samples taken on day 7 after infection (Fig. 3A). Is this an indirect effect, or are CTL numbers in some way dependant on the concurrent presence of tipDCs? It seems that the effect may be local, because it only was evident in the lungs and was not apparent in the regional mediastinal lymph nodes (MLNs) where the virus-specific CD8+ T cell populations were of essentially equivalent magnitudes in CCR2+/+ and CCR2−/− mice (Fig. 3B).
Fig. 3.
tipDCs present antigen to CD8+ T cells in the lungs. (A) CCR2−/− animals have significantly fewer CD8+ T cells specific for the influenza DbPA224–233, KbPB1703–711, and DbPB1-F262–70 epitopes. Results are representative of 2 independent experiments (n ≥ 5) and were analyzed by using the two-tailed Student's t test (P values are indicated). (B) There is no statistically significant difference in the number of antigen-specific CD8+ T cells between CCR2+/+ and CCR2−/− animals in the draining lymph node on day 7 after infection with 105 pfu of x-31. Results are representative of two independent experiments (n ≥ 5) and were analyzed by using the two-tailed Student's t test (P > 0.05 for each comparison). (C) tipDCs isolated from the BAL wash on day 6 after infection present the equivalent of 40 pmol per cell of DbPB1-F262–70 epitope to the DbPB1-F262–70 epitope-specific hybridoma. Bars represent the mean ± SEM of triplicate wells, and the results were analyzed by using the two-tailed Student's t test (P value indicated). (D) tipDCs isolated from the BAL wash of animals infected with the intact x-31 virus (but not the DbNP366–374- and DbPA224–233-disrupted DKO virus) were sufficient to rescue the DbNP366–374 and DbPA224–233 epitope-specific CD8+ T cell response when transferred into the lungs of CCR2−/− animals on day 1 after infection (n = 5). Results were analyzed by using a single-factor ANOVA and Tukey's posthoc test (same letter, P > 0.05; different letters, P < 0.05).
tipDCs Present Antigen to CD8 T Cells in the Lungs.
Because the influenza-specific CD8+ T cell counts were significantly reduced in the lungs (but not the lymph nodes) of CCR2−/− animals (Fig. 3A), we next asked how tipDCs might regulate virus-specific CD8+ T cell numbers. One possibility was that the tipDCs could produce soluble factors that function to support CD8+ T cell survival, division, or both. Therefore, we determined the concentrations of 23 cytokines and chemokines in the cell-free BAL fluid from PR8-challenged CCR2−/− and CCR2+/+ mice and found that there were no significant differences for the key cytokines that are known to be involved in regulating CD8 T cells, including IL-2 and IL-4 (Fig. S5). The next step was to assess the capacity of the tipDCs to function as antigen-presenting cells (APCs). For this experiment, tipDCs (together with neutrophils as a control) were purified from the day 6 BAL wash of PR8-infected B6 mice, then incubated with a CD8+ T cell hybridoma specific for the DbPB1-F262–70 epitope. As expected, the neutrophils failed to stimulate the hybridoma. In contrast, the freshly isolated tipDCs presented the equivalent of 40 pmol per cell of the DbPB1-F262–70 epitope (Fig. 3C). Because no exogenous peptide was added in this experiment, it is clear that the tipDCs had both processed influenza proteins in situ and expressed the PB1-F262–70 peptide/H2Db MHC class I glycoprotein complex on the cell surface while in the virus-infected lung.
Demonstrating a capacity to process and present antigen that can be recognized by a CD8 T cell hybridoma does not establish, however, that tipDCs indeed serve as APCs during influenza virus infection. In fact, it is clear that tipDCs are not the sole APCs driving clonal expansion of influenza-specific CD8+ T cell precursors. We were unable to isolate tipDCs from the draining MLN at any point after infection, and the lymph node CTL responses to the DbPA224–233, KbPB1703–711, and DbPB1-F262–70 epitopes (Fig. 3B) were equivalent for CCR2−/− and CCR2+/+ mice. If tipDCs do serve as APCs in vivo, then this function is manifested outside the draining lymph node.
To further delineate whether tipDCs can function as APCs within the respiratory tract, we purified tipDCs from the BAL of B6 mice that had been infected with either x-31 or a double-knockout (DKO) x-31 virus. The mutated NP366–374 and PA224–233 peptides in the DKO virus do not complex with H2Db, and therefore the responses to these epitopes are lost (21). We then transferred 104 tipDCs recovered from x-31 or DKO-infected mice directly into the airways of CCR2−/− animals that had been infected with x-31 virus i.n. 24 h previously. We found that giving tipDCs from CCR2+/+ animals infected with the intact x-31 virus to CCR2−/− animals was sufficient to recover the DbNP366–374- and DbPA224–233-specific CD8+ CTL responses, whereas those from comparable mice infected with the DKO virus were >10-fold less effective (Fig. 3D). The recovery of the CD8+ T cell response after tipDC transfer was clearly antigen dependent, establishing that tipDCs indeed serve as APCs for CD8+ T cells in the lungs of mice infected with influenza A viruses. These results are in accord with a recent finding that CD8+ T cells and DCs interact in the lungs of influenza-infected mice (22).
Reducing tipDC Accumulation Protects from Lethal Influenza Challenge.
So far we have shown that the number of tipDCs is significantly elevated in mice infected with HP influenza A viruses, that these tipDCs function locally in the respiratory tract as APCs, and that they are required for the full realization of protective CD8+ T cell-mediated immunity. In addition, their complete absence from the lungs of CCR2−/− mice is associated with severe disease, indicating that tipDC ablation is not a viable therapeutic option.
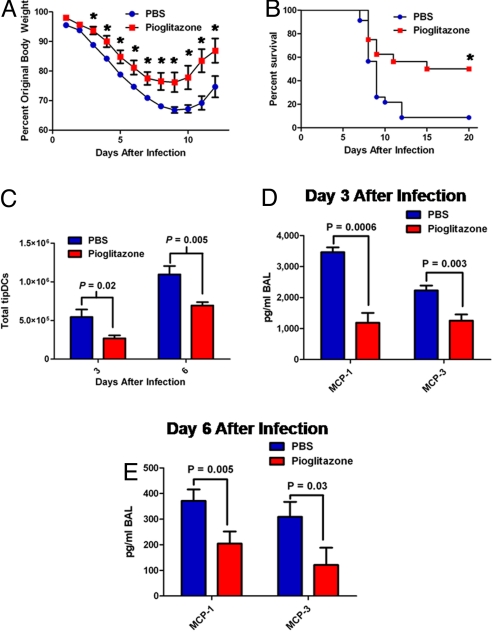
With this in mind, we hypothesized that reducing, without eliminating, tipDC accumulation in the lungs after challenge with HP influenza A viruses may prove beneficial to the host. While searching for pharmacological suppressors of chemokine (C-C motif) ligand 2 (CCL2) secretion, we found published evidence that activation of the peroxisome proliferator-activated receptor-γ (PPAR-γ) with the synthetic agonist pioglitazone reduces the production of a wide range of proinflammatory molecules, including CCL2, TNF-α, and iNOS (23–25). With this knowledge, we speculated that a prophylactic regimen of pioglitazone may provide protection from HP influenza challenge by reducing the number of tipDCs recruited to the airways. With weight loss as a measure for morbidity, B6 mice were treated with either pioglitazone (60 mg/kg in 100 μL of PBS) or PBS (100 μL) via oral gavage daily beginning 3 days before infection with 5 mouse 50% lethal dose (MLD50) of PR8 virus. The pioglitazone-treated animals were less severely affected and recovered more quickly than the PBS controls (Fig. 4A), while mortality was reduced from 92% to 50% (Fig. 4B).
Fig. 4.
Pioglitazone protects from lethal influenza challenge. Prophylactic treatment with pioglitazone significantly decreases host morbidity, presented as mean percentage original body weight ± SEM and representative of 3 independent experiments (n ≥ 5) (interaction effect, P < 0.001; asterisks mark significant simple effects, P < 0.001) (A), increases host survival (Mantel–Cox, P = 0.005) after lethal influenza challenge (103 pfu of PR8) of the pioglitazone-treated group (n = 16) versus PBS-treated controls (n = 23) (B), decreases MCP-1 (CCL2) and MCP-3 accumulation in the BAL wash on days 3 (C) and 6 (D) after infection with 103 pfu of PR8, and decreases tipDC accumulation in the lungs (E). Results are representative of 2 independent experiments (n ≥ 5) and were analyzed with the two-tailed Student's t test (P values are indicated).
Was this protective effect a consequence of decreased inflammation? To investigate this possibility, immune cells within the lungs of mice treated with pioglitazone or PBS and challenged with 5 MLD50 of PR8 virus were enumerated by flow cytometry. Although there was a general decrease in the magnitude of the cellular inflammatory exudate, the sole statistically significant reduction was in tipDC numbers (Fig. 4C). This finding is in accord with a recent study in which increased inflammation via a CCR2-dependent mechanism is associated with increased mortality (26). Furthermore, when we measured the effect of pioglitazone on proinflammatory cytokines and chemokines in the BAL supernatant, only CCL2 (MCP-1) and MCP-3 were reduced to a statistically significant level in the drug-treated animals (Fig. 4 D and E).
To determine whether the protective effects of pioglitazone correlated with the inhibition of virus replication, we measured the virus titers in the BAL fluid from pioglitazone- and PBS-treated animals on days 3, 6, and 9 after infection. No significant differences in virus titers were found at any time point (Fig. S6A). Further, treatment with up to 1 μg/mL pioglitazone (as compared with PBS) did not inhibit virus replication in Madin–Darby canine kidney (MDCK) cells (Fig. S6B). Together, these results indicate that the protection observed in pioglitazone-treated B6 mice reflects the reduced recruitment of tipDCs (via suppression of CCL2 (MCP-1) and MCP-3) rather than any inhibition of virus replication.
Discussion
The present study establishes that the disproportionate increase in lethality for young, healthy individuals infected with HP strains of influenza A viruses correlates with a significant increase in the recruitment of tipDCs to the pneumonic lung. However, although tipDCs promote immune-induced pathology, their complete elimination is detrimental. This reflects that they also function as APCs to facilitate the optimal expansion of effector CD8+ T cells within the virus-infected respiratory tract.
Incidentally, the clear demonstration of increased CD8+ T cell counts induced by the local presence of antigen-presenting tipDCs also provides insight into an old question concerning the nature of T cell-mediated immunity in influenza. It now seems likely that effector CD8+ CTLs do continue to divide at the site of pathology after they leave the lymphoid tissue, although the formal alternative remains (of course) that the tipDCs function to promote antigen-specific CD8+ T cell retention (rather than further cycling) in the infected lung. Either way, the tipDCs act to enhance the protective CD8+ T cell response.
Because CD8+ T cells play a critical role in influenza virus clearance (19, 20), a blunted response compromises control of the infectious process and contributes to morbidity, offsetting any positive consequence that can result from diminished immunopathology. Here, we demonstrate use of the PPAR-γ agonist pioglitazone. We found that prophylactic treatment with pioglitazone is sufficient to reduce morbidity and mortality associated with HP influenza A virus infection. The therapeutic benefit of pioglitazone treatment correlates with diminished levels of CCL2 (MCP-1) and MCP-3 secretion and ultimately reduced numbers of tipDCs recruited to the airways but not with decreased virus replication. What the pioglitazone treatment achieves, in effect, is a better “middle way” between the protective and pathological consequences of immune responsiveness.
The findings from the present analysis thus identify a new therapeutic target for immune manipulation, the tipDCs. Previous studies using global immune suppressants (such as steroids) have failed to protect against lethal influenza virus challenge (27). It is not surprising that such nonspecific immune suppressants confer no advantage, because a systemic reduction in cell-mediated immunity greatly compromises virus clearance. On the basis of the present results, it seems reasonable to suggest that new therapies might be focused on promoting the middle way of reducing but not eliminating the accumulation of tipDCs in the respiratory tract. Moreover, the possibility that targeting tipDC recruitment may be of value for other disorders where an overexuberant immune response promotes pathology and severe disease consequences is worth considering (28).
Methods
Viruses.
The PR8 and x-31 influenza A viruses were obtained from the St. Jude Children's Research Hospital repository. The x-31 DKO virus with mutated, nonresponder DbNP366–374 and DbPA224–233 peptides was generated earlier by using the 8-plasmid reverse-genetics system (21, 29). The HK213 and VN1203 viruses were obtained through the World Health Organization Global Influenza Surveillance Network. Stocks were propagated by allantoic inoculation of 10-day-embryonated chicken eggs, and virus titers were determined as pfus on MDCK monolayers. The experiments with H5N1 viruses were conducted in a U.S. Department of Agriculture-approved BSL 3+ containment facility.
Mice.
The wild-type C57BL/6J (B6) and CCR2−/− mice were purchased from The Jackson Laboratory and were housed under specific-pathogen-free conditions. All mice in this study were used according to protocols approved by the Institutional Animal Care and Use Committee at St. Jude Children's Research Hospital.
Viral Infection and Sample Collection.
Naive mice were anesthetized by i.p. injection with avertin (2,2,2-tribromoethanol) before i.n. challenge with the appropriate dose of virus diluted in 30 μL of sterile, endotoxin-free PBS. Animals were weighed before infection, then monitored daily for weight loss as a measure of morbidity, or euthanized at the appropriate time point for sample collection. BAL fluid was collected in three 1-mL washes with HBSS. Cells were pelleted by light centrifugation, and total numbers per BAL sample were determined by using a Coulter counter (IG Instrumenten).
Flow-Cytometric Analysis.
The BAL cells were stained with the appropriate combination of FITC-labeled anti-Ly6g (1A8; BD Biosciences), phycoerythrin (PE)-labeled anti-MHC class II (AF6–120.1; BD Biosciences), PE-Cy5-labeled anti-CD11c (N418; eBioscience), Alexa Fluor 647-labeled anti-Ly6c (ER-MP20; AbD Serotech), allophycocyanin (APC)-Cy7-labeled anti-CD11b (M1/70; BD Biosciences), FITC-labeled anti-CD4 (RM4.5; BD Biosciences), PE-labeled anti-NK1.1 (PK136; BD Biosciences), APC-labeled αβT cell receptor (H57–597; BD), APC-Cy7-labeled anti-CD8 (53–6.7; eBioscience), or PE-labeled anti-TNF-α (MP6-XT22; BD Biosciences) after blocking of the Fc receptor with anti-CD32/CD16 (BD Biosciences) at 4 °C. For CD8+ T cell analysis, cells were stained with tetramers specific for the DbPA224–233, KbPB1703–711, or DbPB1-F262–70 epitopes for 1 h at room temperature before staining with the monoclonal antibodies (mAbs). Intracellular staining was performed by staining the cells for surface markers before fixing in 2% paraformaldehyde and permeabilizing with Perm/Wash buffer (BD Biosciences). For iNOS staining, cells were incubated with goat anti-iNOS (M-19; Santa Cruz Biotechnology) followed by FITC-conjugated anti-goat IgG (Jackson ImmunoResearch). Data were acquired by using either a FACSCalibur or FACScan (BD Biosciences) and analyzed by using FloJo (Tree Star). For characterization of cell types, a large gate was drawn to include the monocyte and lymphocyte populations from forward scatter vs. side scatter. Cells were further differentiated by expression of cell-specific markers as indicated in Table S1. The number of viable cells per BAL sample was determined by using a Coulter counter (IG Instrumenten), and individual cell subsets were calculated by multiplying the percentage of each cell type (as determined by FACS) by the total number of viable cells recovered per BAL.
Cytokine and Chemokine Quantification.
Mouse cytokines and chemokines in the cell-free BAL fluid were measured by using the Bio-Rad multiplex assay.
Virus Titer Determination.
Cell-free BAL fluid samples were titered by plaque assay on MDCK cells. Near-confluent 25-cm2 MDCK cell monolayers were infected with 1 mL of six 10-fold dilutions of BAL fluid for 1 h at 37 °C, then washed with PBS before adding 3 mL of MEM containing 1 mg/mL l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (Worthington) and 0.9% agarose. Cultures were incubated at 37 °C, 5% CO2 for 72 h. Plaques were visualized with crystal violet.
T Cell Hybridoma Stimulation by tipDCs.
Wild-type B6 mice were infected with 105 pfu of x-31. On day 6 after infection, BAL fluid was collected, and tipDCs were purified by cell sorting (MoFlo; Beckman Coulter). The tipDCs (≈1 × 104) were cocultured in 96-well plates with 2 × 104 DbPB1-F262-specific hybridoma cells (line F2-54) for 24 h. The F2-54 hybridoma line reacts positively by IL-2 production with the DbPB1-F262 epitope but negatively with DbPA224, KbPB1703, KbNS2114, and DbNP366. At the same time, a standard curve was generated by pulsing uninfected splenocytes with serially diluted PB1-F262 peptide, then coculturing with the F2-54 hybridoma. IL-2 production, which in this assay correlates directly to the MHC class I expression on the APC cell surface, was measured by ELISA using purified anti-IL-2 and biotin–anti-IL-2 antibodies (BD BiosciencesBD) following the manufacturer's directions. The concentration of epitope was calculated based on IL-2 production derived from the standard curve, and the amount of peptide presented per APC was calculated by dividing the determined amount of peptide presented by the number of APCs added to the assay.
tipDC Transfer.
The B6 mice were infected i.n. with 105 pfu of either the intact x-31 or the DKO virus with disrupted DbNP366–374 and DbPA224–233 epitopes. On day 6 after infection, tipDCs were isolated from the BAL fluid by using a MoFlo (Beckman Coulter) to >95% purity. The CCR2−/− animals were infected i.n. with 105 pfu of x-31. On day 1 after infection, 104 tipDCs purified from animals given either x-31 or DKO were transferred intratracheally into CCR2−/− animals. Six days after tipDC transfer (day 7 after infection) BAL fluid was collected, and cells were stained with tetramers specific for the DbNP366–374 and DbPA224–233 epitopes at room temperature for 1 h. After blocking the Fc receptor with anti-CD32/CD16 (BE Biosciences) at 4 °C, the cells were then stained with APC-Cy7-labeled anti-CD8 (eBioscience) and analyzed by flow cytometry (FACScan, BD Biosciences Pharmingen). Total cell numbers per BAL sample were determined by using a Coulter counter (IG Instrumenten).
Pioglitazone Treatment.
The B6 mice were treated with 60 mg/kg of pioglitazone (suspended in 100 μL of PBS) via oral gavage beginning on day 3 before infection and continued daily thereafter. The control mice were given 100 μL of PBS, and both groups were challenged with 5 MLD50 of PR8. Weight loss was monitored daily as a measure of morbidity. On days 3 and 6 after infection, groups of mice from the treatment and control groups were euthanized. The BAL fluid was collected in three 1-mL washes with HBSS, and cells were isolated by light centrifugation. The total cell count per BAL was determined by using a Coulter counter, and the cells were analyzed by flow cytometry. Virus titers in BAL fluid were determined by plaque assay. Cytokines and chemokines were quantified by using the Bio-Rad multiplex assay, except for MCP-3, which was measured by ELISA (Bender MedSystems).
Statistical Analysis.
Data are presented as mean ± SEM. Time course data, i.e., cell counts and cytokine and chemokine expression, were analyzed with multiple regression. When there was a significant interaction effect, the simple effect of virus or treatment was investigated by performing ANOVA for each time point. This analysis accounted for multiple comparisons by using the mean-square error from the regression analysis to calculate the F statistic. If the interaction effect was not significant, then model simplification was performed to find the minimal adequate model. Similar techniques were used to analyze weight loss after infection. However, because each mouse was measured at every time point, mouse was included as a random effect in the regression model. Model simplification was performed in R 2.8.0 (www.R-project.org). All remaining analyses were performed in JMP 4.0.4 (SAS Institute).
Supplementary Material
Acknowledgments.
We thank Hui-Ling Yen and Jacco Boon for advice; Richard Webby for the DKO virus; Scott Krauss, Melissa Morris, Richard Cross, James Knowles, Jennifer Rogers, Ashley Webb, Yolanda Griffin, and Cedric Proctor for excellent technical assistance; Ray Kuhn and Sharon Naron for editorial assistance; Julie Groff for assistance with Fig. S1; and the World Health Organization Global Influenza Surveillance Network for providing the H5N1 viruses. The authors dedicate this manuscript to the memory of our good friend and colleague, the late Cedric Proctor. Cedric's support in our high-containment facility was critical for completion of the present study. This work was supported by Contract HHSN266200700005C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, and by the American Lebanese Syrian Associated Charities.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 4961.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900655106/DCSupplemental.
References
- 1.Meltzer MI, Cox NJ, Fukuda K. The economic impact of pandemic influenza in the United States: Priorities for intervention. Emerg Infect Dis. 1999;5:659–671. doi: 10.3201/eid0505.990507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horimoto T, Kawaoka Y. Influenza: Lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 3.Loo YM, Gale M., Jr Influenza: Fatal immunity and the 1918 virus. Nature. 2007;445:267–268. doi: 10.1038/445267a. [DOI] [PubMed] [Google Scholar]
- 4.Palese P. Influenza: Old and new threats. Nat Med. 2004;10:S82–S87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 5.Smith K. Concern as revived 1918 flu virus kills monkeys. Nature. 2007;445:237. doi: 10.1038/445237a. [DOI] [PubMed] [Google Scholar]
- 6.Taubenberger JK, et al. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 7.Tumpey TM, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 8.Taubenberger JK, Morens DM. 1918 Influenza: The mother of all pandemics. Emerg Infect Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO (World Health Organization, Geneva, Switzerland) 2009. [Accessed January 23, 2009]. Available at www.who.int/csr/disease/avian_influenza/country/cases_table_2009_01_22/en/index.html.
- 10.Kash JC, et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobasa D, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 12.Kobasa D, et al. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature. 2004;431:703–707. doi: 10.1038/nature02951. [DOI] [PubMed] [Google Scholar]
- 13.Chan MC, et al. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir Res. 2005;6:135. doi: 10.1186/1465-9921-6-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung CY, et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: A mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–1837. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 15.Serbina NV, et al. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- 16.Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol. 2006;7:311–317. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- 17.Dawson TC, et al. Contrasting effects of CCR5 and CCR2 deficiency in the pulmonary inflammatory response to influenza A virus. Am J Pathol. 2000;156:1951–1959. doi: 10.1016/S0002-9440(10)65068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin KL, et al. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J Immunol. 2008;180:2562–2572. doi: 10.4049/jimmunol.180.4.2562. [DOI] [PubMed] [Google Scholar]
- 19.Doherty PC, et al. Effector CD4+ and CD8+ T-cell mechanisms in the control of respiratory virus infections. Immunol Rev. 1997;159:105–117. doi: 10.1111/j.1600-065x.1997.tb01010.x. [DOI] [PubMed] [Google Scholar]
- 20.Topham DJ, Tripp RA, Doherty PC. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J Immunol. 1997;159:5197–5200. [PubMed] [Google Scholar]
- 21.Webby RJ, et al. Protection and compensation in the influenza virus-specific CD8+ T cell response. Proc Natl Acad Sci USA. 2003;100:7235–7240. doi: 10.1073/pnas.1232449100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGill J, Van Rooijen N, Legge KL. Protective influenza-specific CD8 T cell responses require interactions with dendritic cells in the lungs. J Exp Med. 2008;205:1635–1646. doi: 10.1084/jem.20080314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Gregorio GB, et al. Expression of CD68 and macrophage chemoattractant protein-1 genes in human adipose and muscle tissues: Association with cytokine expression, insulin resistance, and reduction by pioglitazone. Diabetes. 2005;54:2305–2313. doi: 10.2337/diabetes.54.8.2305. [DOI] [PubMed] [Google Scholar]
- 24.Haraguchi G, et al. Pioglitazone reduces systematic inflammation and improves mortality in apolipoprotein E knockout mice with sepsis. Intensive Care Med. 2008;34:1304–1312. doi: 10.1007/s00134-008-1024-9. [DOI] [PubMed] [Google Scholar]
- 25.Ito H, Nakano A, Kinoshita M, Matsumori A. Pioglitazone, a peroxisome proliferator-activated receptor-γ agonist, attenuates myocardial ischemia/reperfusion injury in a rat model. Lab Invest. 2003;83:1715–1721. doi: 10.1097/01.lab.0000106724.29121.da. [DOI] [PubMed] [Google Scholar]
- 26.Herold S, et al. Lung epithelial apoptosis in influenza virus pneumonia: The role of macrophage-expressed TNF-related apoptosis-inducing ligand. J Exp Med. 2008;205:3065–3077. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salomon R, Hoffmann E, Webster RG. Inhibition of the cytokine response does not protect against lethal H5N1 influenza infection. Proc Natl Acad Sci USA. 2007;104:12479–12481. doi: 10.1073/pnas.0705289104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowes MA, et al. Increase in TNF-α and inducible nitric oxide synthase-expressing dendritic cells in psoriasis and reduction with efalizumab (anti-CD11a) Proc Natl Acad Sci USA. 2005;102:19057–19062. doi: 10.1073/pnas.0509736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann E, et al. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.