Figure 4. DMD refinement of RNA secondary and tertiary structure.
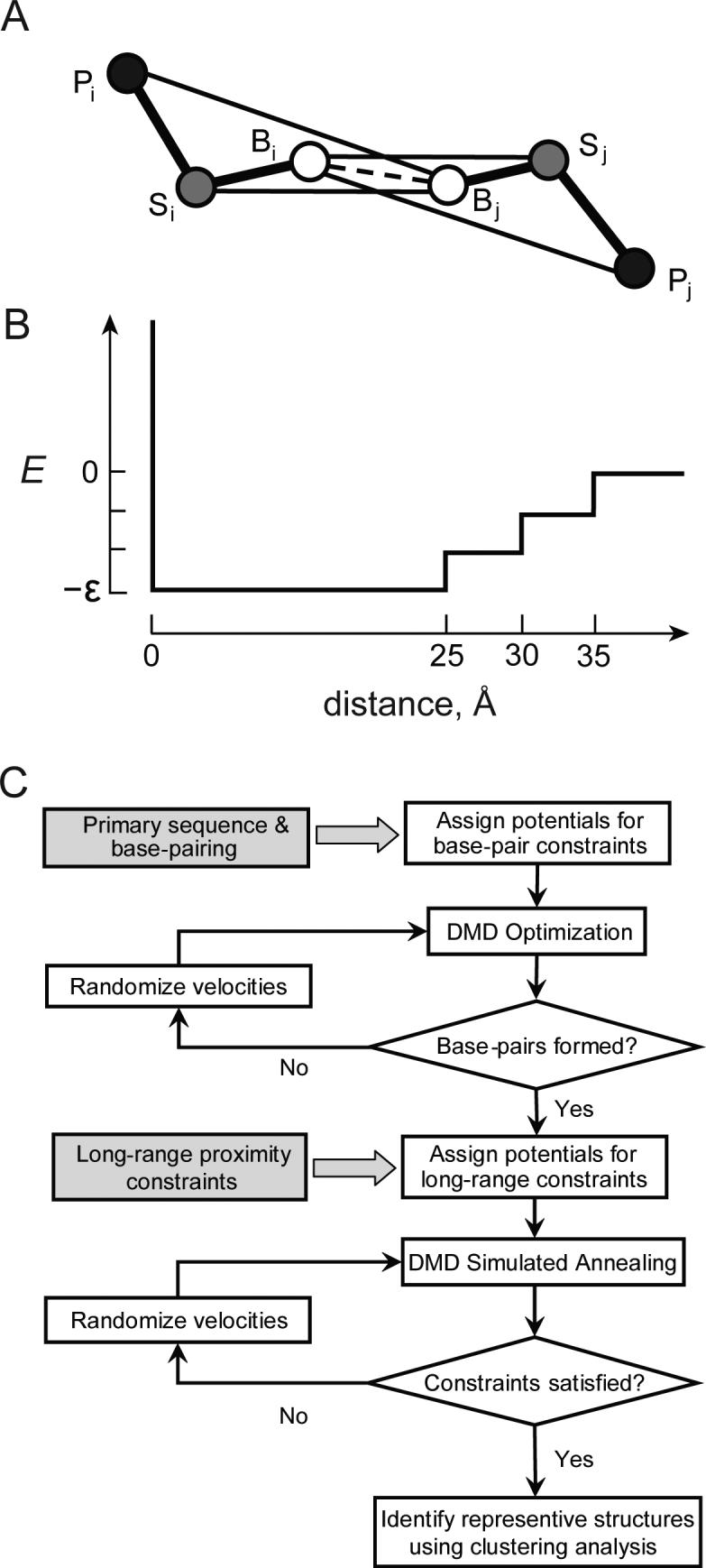
(A) Three pseudoatom model in which an RNA nucleotide is represented by base (B), ribose (S) and phosphate (P) groups. Lines indicate pair-wise potentials between atoms: Bi-Bj, base pairing interactions, Bi(j)-Sj(i) interaction energies for inter-nucleotide orientation dependences. The Bi(j)-Pj(i) interaction between base and phosphate reflects rigidity of RNA helices. (B) Energy diagram for the long-range constraint potential. (C) Algorithm for DMD refinement.
