Figure 5.
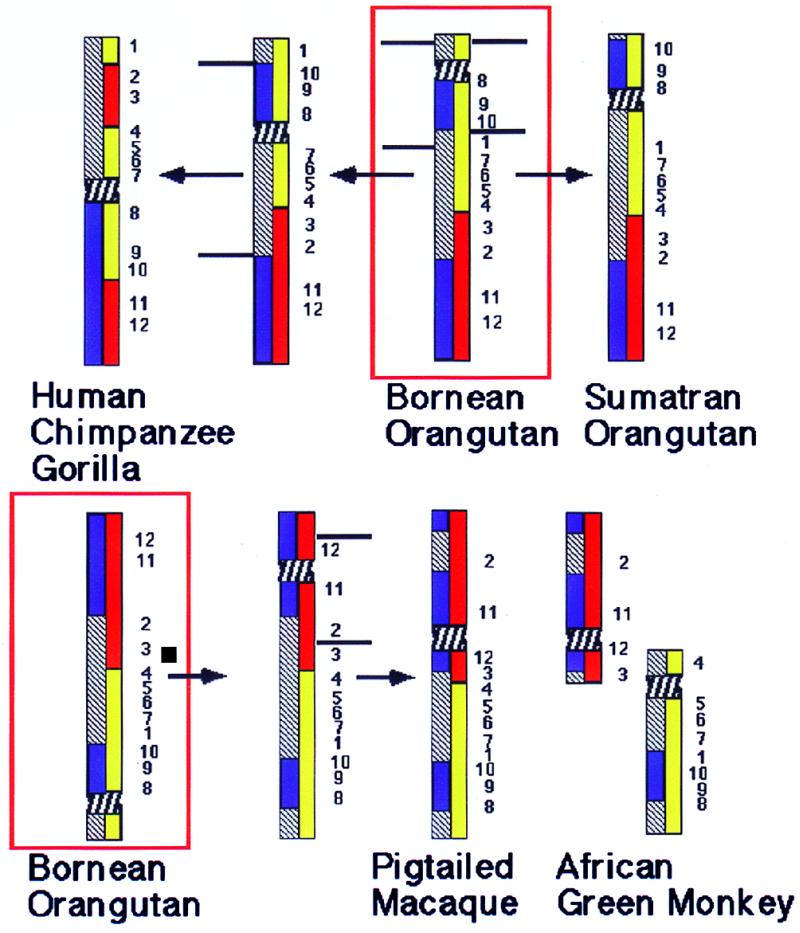
The most parsimonious reconstruction of chromosome 3 rearrangements in higher primates. Blue bars within a chromosome represent the hybridization signals obtained with a painting probe specific for human chromosome 3q. Yellow and red bars show the hybridization signals obtained with African green monkey chromosome 15 and 22 paints, respectively. The horizontal lines show inversion break points and the arrows the direction of changes. Numbers indicate the location and order of YAC signals. The red boxes frame the assumed ancestral condition for all higher Old World primates as found in the Bornean orangutan. (Upper) To derive the homologs of human/African great apes from that of Bornean orangutan two overlapping inversions are necessary. First YACs 8–10 and 1 would have to be inverted, leading to an intermediate chromosome type. In a second inversion a fragment including YACs 2–7, the centromere, and YACs 8–10 were inverted. The signal derived from African green monkey chromosome paints are split in two segments each. The Sumatran orangutan homolog can be derived from that of the Bornean orangutan by a single pericentric inversion involving breakpoints between YACs 1 and 10 and the telomere. (Lower) A centromere inactivation/activation and a single pericentric inversion is needed to derive the ancestral Old World monkey homolog as seen in both the macaque and snub-nosed monkeys from that of the Bornean orangutan. For better comparison the orangutan homolog has been inverted. C. aethiops homologs can be derived from the ancestral Old World monkey homolog by a fission between YACs 3 and 4 and the activation of a new centromere.
