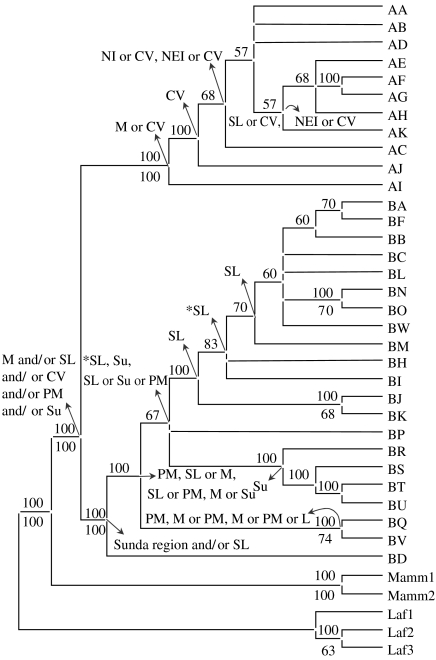
Figure 2.
Consensus MP tree of Asian elephant haplotypes, rooted with African elephant (Laf1, Laf2 and Laf3) and woolly mammoth (Mamm1 and Mamm2) haplotypes as outgroups. Consensus values are given above the branch and the bootstrap values (based on 1000 replicates) above 50 per cent are given below the branch adjoining the corresponding node. Ancestral areas inferred from dispersal–vicariance analysis are shown with arrows from the corresponding nodes. Abbreviations used: B, Borneo; CV, Cambodia–Vietnam; L, Laos; M, Myanmar; NEI, northeastern India and Bhutan; NI, northern India; PM, Peninsular Malaysia; SI, southern India; SL, Sri Lanka; Su, Sumatra. Asterisks indicate results based on shown as well as alternative arrangements at those nodes.