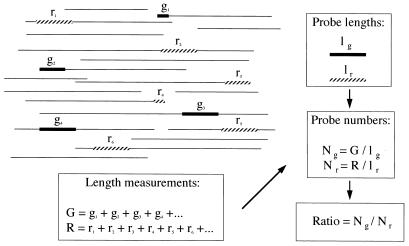
Figure 2.
Experimental scheme. Dashed boxes (ri) represent labeled probes hybridized to fragments of genomic DNA and detected by Texas red fluorescence (r, red detection; index i, rank of the observed fragment). These probes are used to count the number of genomes present on the coverslip. This number is given by the sum of the lengths of the corresponding hybridization signals divided by the known length of the region (Nr = R/lr; lr = normal length of region). The black boxes (gi) represent linear FITC fluorescent signals specifically hybridized to the region whose copy number is to be determined (g, green detection; index i, rank of the observed fragment). Again, it is the total length of the measured hybridization signals divided by the known size of the region that yields its respective copy number per coverslip (Ng = G/lg). The copy number of the region present in the sample of genomic DNA then is obtained by dividing the number of target regions present on the coverslip by the number of genomes scanned. The boxes are shown to be of varying sizes to emphasize that detected signals often are truncated because of shearing of the genomic DNA during its preparation.