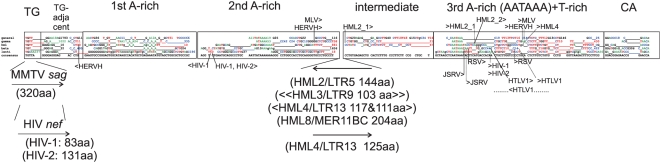
Figure 6. Combined alignment of Viterbi consensus sequences (“SuperViterbi” alignment) from the five HMMs.
Each Viterbi is represented as a match state consensus sequence with the most common nucleotide, disregarding absence of fit. The SuperViterbi alignment was based on a Viterbi HMM alignment of the five Viterbi consensuses (Fig S7). In this secondary HMM, most primary match states were retained as match states (upper case). A few became insert states (lower case). Insert state figures from each Viterbi consensus were then added. Insert state numbers above 20 were given an additional space to denote a continuity break. Insert states occurred nonrandomly. Taking them, together with conspicuous and previously known motifs like TG, TATA, AATAAA, T-rich and CA, into account, modules could be discerned after conservative manual sliding in the original SuperViterbi alignment. The origins of the HMMs are further clarified in Table 1. A consensus based on the manually adjusted SuperViterbi alignment is shown below the other sequences. Landmarks derived from well characterized LTRs were plotted in the manually adjusted SuperViterbi alignment. Transcriptional start sites are shown as full lines, where viral abbreviations were followed by “>” if in sense and by “<” if in antisense direction. Polyadenylation sites were shown by dotted lines, with viral abbreviation preceded by “>”. Abbreviations and accession numbers: TSS and polyadenylation sites are shown for HIV-1 (strain hxb2), HIV-2 (D00835), HML2_1 [79] , or alternatively HML2_2 [34] , HML4 [27], [55], JSRV (AF357971), HERV-H [23], MLV (J01998), RSV (NC_001407) and HTLV-1 (HL1PROP). TSSs and polyadenylation sites were mapped onto the beta HMM consensus except HERV-H and MLV which were mapped on the gamma HMM consensus. Positions of ORFs are also given. HIV-1 nef (MNCG) reaches into around half of U3. The HML ORFs, found in RepBase or Clustal LTR consensus sequences, are either contained between the second A-rich and intermediate modules, or overlap with several modules (HML3). They are antisense to the standard retroviral transcriptional sense, except for one (HML4). The module-overlapping HML3 ORF is symbolized with “<<..>>”.