Figure 2.
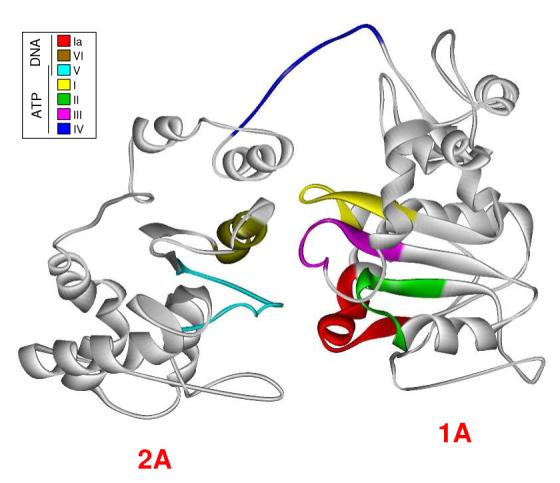
Homology model of Dda. The crystal structure of RecD was replaced residue for residue with its corresponding amino acid residue from its alignment with Dda. The energy of the structure was minimized periodically during the process. Helicase motifs are shown in the inset box and are grouped by their conserved functions. Motifs known to interact with ATP/ADP are located in the cleft between RecA-like domains of the protein while motifs associated with ssDNA interaction are located on the bottom face of the protein. The exception is Motif IV which has been reported to be involved in ssDNA interaction but is located near the ATP binding pocket in our model.
