Figure 2.
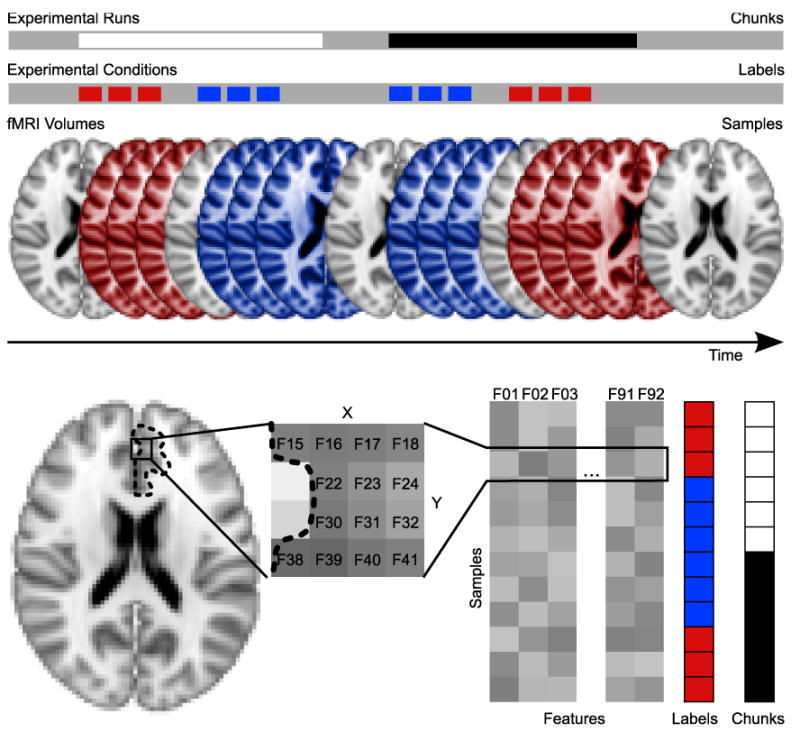
Terminology for classifier-based analyses in PyMVPA. The upper part shows a simple block-design experiment with two experimental conditions (red and blue) and two experimental runs (black and white). Experimental runs are referred to as independent chunks of data and fMRI volumes recorded in certain experimental conditions are data samples with the corresponding condition labels attached to them (for the purpose of visualization the axial slices are taken from the MNI152 template downsampled to 3 mm isotopic resolution). The lower part shows an example ROI analysis of that paradigm. All voxels in the defined ROI are considered as features. The three-dimensional data samples are transformed into a two-dimensional samples×feature representation, where each row (sample) of the data matrix is associated with a certain label and data chunk.
