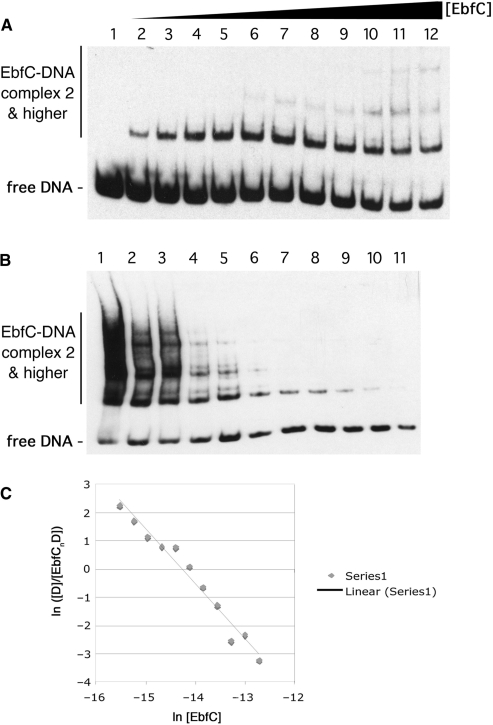
Figure 4.
Determination of the Kd of EbfC binding to wild-type erpAB Operator 2 DNA. (A) Titration of labeled probe b-WT, a 124 bp segment including erpAB Operator 2 (Figure 1), with increasing concentrations of purified, recombinant EbfC. Each lane represents equally loaded reactions consisting of 100 pM b-WT plus 0, 68, 140, 210, 270, 340, 410, 480, 550, 620, 680 or 820 nM EbfC for lanes 1–12, respectively. (B) Serial dilution of non-equilibrated EbfC + DNA reactions. A mixture of 3 μM EbfC and 4 nM b-WT was produced and immediately serially diluted with three volumes of the previous tube and one volume of binding buffer. Final concentrations of EbfC were 3.0, 2.3, 1.7, 1.3, 0.98, 0.74, 0.56, 0.42, 0.32, 0.24, 0.18 μM and concentrations of probe b-WT were 4.0, 3.1, 2.4, 1.9, 1.4, 1.1, 0.88, 0.68, 0.52, 0.41, 0.31 nM for the mixtures in lanes 1–11, respectively. These reactions were allowed to equilibrate before loading equal volumes onto the gel to perform the mobility shift assay. (C) Band intensities from (B) were determined and ln([D]/[EbfCnD]) was graphed as a function of ln [EbfC]. The slope of the resulting line is equal to −n, the negative value of the binding stoichiometry and the x-intercept is the point at which n ln [EbfC] = lnKd.