Abstract
The DNA structure-selective endonuclease Mus81-Mms4/Eme1 incises a number of nicked joint molecule substrates in vitro. 3′-flaps are an excellent in vitro substrate for Mus81-Mms4/Eme1. Mutants in MUS81 are synthetically lethal with mutations in the 5′-flap endonuclease FEN1/Rad27 in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Considering the possibility for isoenergetic interconversion between 3′- and 5′- flaps, these data are consistent with the hypothesis that Mus81-Mms4/Eme1 acts on 3′-flaps in vivo. FEN1/Rad27 prefers dually flapped substrates and cleaves in a way that allows direct ligation of the resulting nick in the product duplex. Here we test the activity of Mus81-Mms4 on dually flapped substrates and find that in contrast to FEN1/Rad27, Mus81-Mms4 activity is impaired on such substrates, resulting in cleavage products that do not allow direct religation. We conclude that Mus81-Mms4, unlike FEN1/Rad27, does not prefer dually flapped substrates and is unlikely to function as a 3′-flapase counterpart to the 5′-flapase activity of FEN1/Rad27. We further find that joint molecule incision by Mus81-Mms4 occurs in a fashion determined by the branch point, regardless of the position of an upstream duplex end. These findings underscore the significance of a nick adjacent to a branch point for Mus81-Mms4 incision.
INTRODUCTION
Mus81-Mms4/Eme1 functions in a subset of eukaryotic recombination pathways, where its nuclease activity is applied to replication fork recovery during S-phase and crossover formation between homologous chromosomes in meiosis (1–5). Mus81 and Mms4/Eme1 are paralogs, evolutionarily related by gene duplication and divergence events that occurred at least 2.5 billion years ago, the projected limit for the origin of eukaryotes. Their ancestral genes, however, are arguably older, present in archaeal genomes as XPF (Crenarchaea) or Hef (Euryarchaea) (6). Euryarchaeal Hef is a multifunctional polypeptide comprising helicase/translocase and endonuclease domains, and its coupled activities may represent a paradigm for understanding the activities of all XPF-related enzymes. The helicase/translocase and endonuclease domains are functionally associated in Hef but have been separated by mutational inactivation of the helicase/translocase domain in Crenarchaeal and eukaryotic XPFs. In eukaryotes, at least four XPF relatives exist as two functional pairs. Two paralogs retain a conserved nuclease domain (XPF and Mus81), whereas two other paralogs are characterized by a degenerate nuclease domain (ERCC1 and Mms4/Eme1). Each ‘active’ paralog is paired with an ‘inactive’ paralog, making the XPF family a set of asymmetric heterodimers in eukaryotes. In vertebrates, the XPF paralog set includes an additional pair, FAAP24-FANCM (7). Neither FAAP24 nor FANCM is characterized as an active nuclease, and instead the FAAP24-FANCM complex uses ATP hydrolysis to move on DNA, an activity presumably mediated by its helicase/translocase domain. Eukaryotic XPF paralogs therefore compose a small family of nuclease and helicase/translocase functions variably retained, lost or mutationally inactivated among the XPF-ERCC1, Mus81-Mms4/Eme1 and FAAP24-FANCM representatives.
Despite the various permutations of active helicase/translocase and nuclease domains among XPF paralogs, one commonality among all XPF family members across archaea and eukaryotes is their binding to jointed structures in DNA. XPF-ERCC1 and their orthologs recognize bubble structures and 3′-flaps in nucleotide excision repair and single-strand annealing, and function in the repair of large insertion/deletion loops in heteroduplex DNA during meiotic recombination; Mus81-Mms4/Eme1 and their orthologs recognize D-loops, replication forks, nicked Holliday junctions and 3′-flaps in eukaryotic replication fork recovery and meiosis; FAAP24-FANCM recognize model replication forks and Holliday junctions in eukaryotic replication fork recovery and interstrand crosslink repair (1,4,5,8). However, the mechanisms by which the eukaryotic XPF paralogs with nuclease function (XPF-ERCC1, Mus81-Mms4/Eme1) identify sanctioned DNA joint molecule substrates in vivo and catalyze the hydrolysis of a phosphodiester bond in duplex DNA remain unclear. Structural implications from Crenarchaeal XPF make a compelling case that bending of DNA arms in the vicinity of a branch point may underpin conformational changes that position the nuclease domain at its target phosphodiester bond (9). If XPF endonucleases use DNA bending during their substrate-sampling mode, they may be aided in vivo by associated proteins that use ATP hydrolysis to impart energetically unfavorable structural features to DNA—including structural properties required for optimal nuclease processing. The recently solved crystal structure of a truncated zebra fish–human Mus81-Eme1 hybrid complex and its accompanying mutational analysis is consistent with the bending of substrate junction arms by binding to surfaces along the protein (10).
An important question for all XPF paralogs concerns the challenge of targeting a phosphodiester bond for hydrolysis in only specific DNA joint molecules, and not indiscriminately in any joint molecule that appears reasonably compatible in vitro. In other words, how are a number of potential in vitro substrates limited to a subset of realized substrates in vivo? This question is of particular concern for Mus81-Mms4/Eme1, the XPF endonuclease most clearly associated with eukaryotic recombination subpathways. A number of joint molecule intermediates can be envisioned in models of DNA repair and recombination. The substrate selectivity of Mus81-Mms4/Eme1 in vitro makes it difficult to assign an in vivo joint molecule target (1–5). DNA substrates that include 3′-flapped joint molecules, displacement loops (D-loops), replication forks and nicked Holliday junctions can be genetically and biochemically upheld as potential substrates (11–18). Holliday junctions covalently sealed in every DNA strand at the junction branch point also remain candidate substrates in vivo, advocated by biochemical studies of the fission yeast and human protein complexes (19–21), by electron micrographs from fission yeast meiosis that suggest the accumulation of single Holliday junctions in the absence of Mus81 (22) and by in vivo studies using plasmid-borne extruded cruciforms in Saccharomyces cerevisiae (23). Most preparations of Mus81-Mms4 are not active on Holliday junctions in vitro, however (11,15,16). We recently reported that S. cerevisiae Mus81-Mms4 isolated to apparent homogeneity from its native eukaryotic host in the absence or presence of genotoxic stress is not active on Holliday junctions in isolation (13). Instead, all substrates that are incised with any proficiency by the enzyme in vitro have a DNA backbone discontinuity (nick) inherent to the substrate core branch point.
We therefore undertook a biochemical approach to further understand properties of the junction branch point that are important to Mus81-Mms4 substrate processing. Specifically, we queried the effect of modifications to the junction branch point at the position of a backbone nick on turnover, binding and incision position. Mus81-Mms4 has been suggested to function as a 3′-flap endonuclease in vivo (2,5,24–26). This model is supported by the Mus81-Mms4/Eme1 biochemistry, as 3′-flaps were determined to be among the substrates most efficiently processed by Mus81-Mms4 in vitro (13–15,25,27). The 3′-flapase model also rationalizes the synthetic lethality of mus81 and rad27 (encoding the FEN1 5′-flapase) mutations in S. cerevisiae and Schizosaccharomyces pombe in vivo (25,28). 3′-flaps and 5′-flaps are envisioned to exist in an isoenergetic equilibrium, requiring the presence of a 3′ or 5′ flapase but not tolerating elimination of both (2).
FEN1/Rad27 is an archaeal/eukaryotic 5′-flap endonuclease that preferentially incises dually flapped substrates over substrates with 5′-flaps alone, generating nicked duplex products (29–32). DNA-strand-displacement synthesis during Okazaki fragment processing relegates RNA primers to 5′-flaps, which are removed by FEN1/Rad27. The endonuclease also functions in long-patch base excision repair, in which a damaged nucleotide is similarly consigned to a 5′-flap that can be cut by a 5′-flap endonuclease. In both cases, the 5′-flap cut by FEN1/Rad27 is generated during DNA-strand-displacement synthesis, and the joint molecule most likely targeted by the nuclease is a dually flapped intermediate in which a single-nucleotide 3′-flap abuts a longer 5′-flap. Hydrolysis of the phosphodiester bond between the first and second bases 3′ to the 5′-flap generates a ligatable nick when the single-nucleotide 3′-flap anneals, following 5′-flap removal. Biochemical observations showing dually flapped substrate preference were subsequently supported by structural evidence, showing that a 3′-flap-binding pocket is conserved in FEN1 homologs in archaea and eukaryotes (33,34).
By analogy to FEN1/Rad27, a 3′-flap endonuclease might also interact with a dually flapped substrate that arises during dynamic, isoenergetic interconversion of 3′- and 5′-ssDNA flaps. Mus81-Mms4 generates nonligatable, gapped products on substrates with 3′-flaps alone and has not been tested on dually flapped products (25). We performed a kinetic analysis to determine KM and kcat for S. cerevisiae Mus81-Mms4 on 3′-flap and variant substrates. We demonstrate that dually flapped substrates and gapped substrates impair catalysis by Mus81-Mms4. In particular, dually flapped substrates reduce catalytic turnover by Mus81-Mms4 and the products remain nonligatable, in contrast to FEN1/Rad27. Furthermore, our results indicate that Mus81-Mms4 incises primarily four nucleotides 5′ to a junction branch point, and gauges the position of incision by reference to the branch point and not to upstream duplex end position.
EXPERIMENTAL PROCEDURES
Purification of His10-FLAG-Mus81/GST-Mms4 by sequential affinity selection for fusion tag
Assays shown in Figures 1–6 were performed with heterodimer purified as described by Ehmsen and Heyer (13).
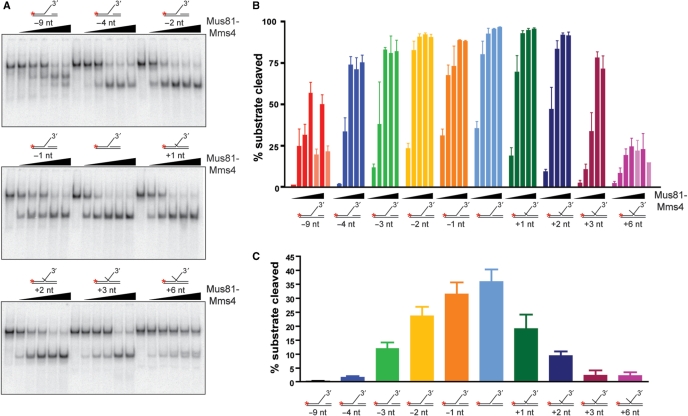
Figure 1.
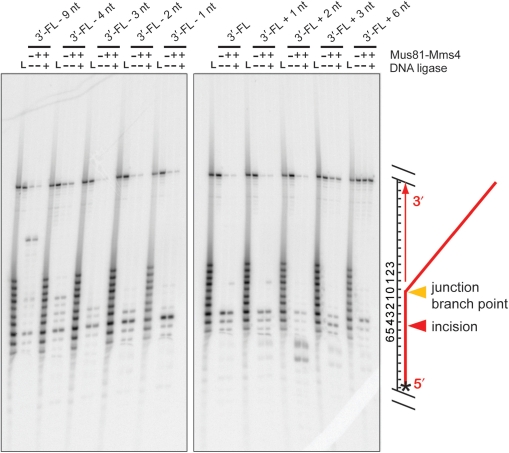
Mus81-Mms4 prefers duplex DNA flush to a joint molecule branch point. (A) Sample gels. Mus81-Mms4 activity was tested on DNA joint molecules related to a 3′-flapped structure, varied only by the 5′ end position of upstream duplex DNA. Duplex arm position was retreated from the branch point by 9 nt, 4 nt, 3 nt, 2 nt and 1 nt, and advanced from the branch point to a 5′ flap by 1 nt, 2 nt, 3 nt and 6 nt. Substrate concentration was defined by nonradiolabeled joint molecule fixed at 50 nM, and heterodimer was titrated at 0, 5, 10, 20, 50 and 100 nM heterodimer. Reactions were incubated at 30°C for 30 min and resolved by electrophoresis on native 10% PAGE–TBE gels, 100 V, 65 min. (B) Quantitation of results in (A), expressed as percent substrate cleaved during the nuclease assay. Incision at alternative sites occurs at high heterodimer concentrations (50 and 100 nM heterodimer) on structures with a 9-nt gap or 6-nt 5′-flap, indicated on the graph by pale bars. (C) Results in (A) and (B) under assay conditions where Mus81-Mms4 is limiting relative to substrate (5 nM heterodimer). The means and standard error of three independent assays are plotted.
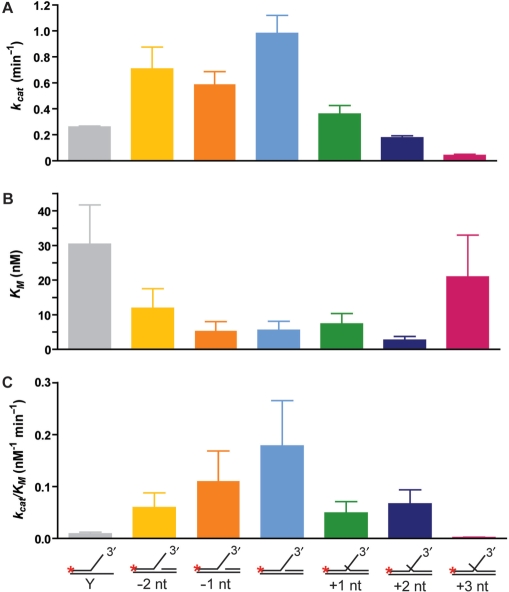
Figure 2.
Comparison of Mus81-Mms4 kinetic values in relation to DNA advanced or retreated from the branch point. Kinetic parameters on DNA joint molecules related to a 3′-flapped substrate are plotted in direct comparison to the 3′-flapped structure. Kinetic analysis was performed on structures with the upstream 5′ duplex position retreated from the branch point by 1 nt (–1 nt) and 2 nt (–2 nt), or with the upstream duplex absent (Y); and on structures with the upstream 5′ duplex position advanced from the branch point to form a short 5′ flap of 1 nt (+1 nt), 2 nt (+2 nt) or 3 nt (+3 nt). (A) kcat (nM min−1), (B) KM (nM), (C) selectivity coefficient kcat/KM (nM−1 min−1). The data for the Y substrate are from Ehmsen and Heyer (13).
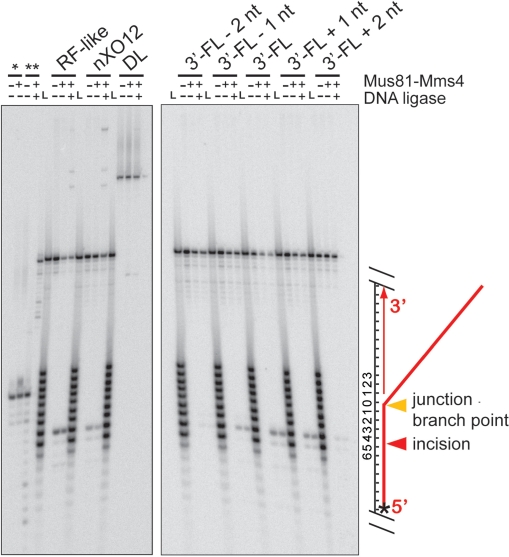
Figure 3.
Mus81-Mms4 incises joint molecules at the branch point, adjacent to a phosphodiester backbone discontinuity. Denaturing urea–PAGE analysis of substrate incision sites on RF-like, nXO12, D-loop (DL) and five 3′-FL-related structures. Assays were performed with 10 nM His10-FLAG-Mus81/GST-Mms4, 50 nM substrate, 30 min at 30°C. Where indicated, reactions were then supplemented with 0.5 mM ATP/3 mM Mg(OAc)2 and 10 U T4 DNA ligase, with incubation at room temperature for 15 min. All assays were terminated by boiling at 95°C, 2 min, with immediate transfer to ice. Nicked duplex DNA without a 5′-phosphate (*) is unligated by T4 DNA ligase, whereas nicked duplex DNA with a 5′ phosphate (**) is ligated by T4 DNA ligase. ‘L’ represents an oligonucleotide size ladder. The scheme on the right side of the gel illustrates the substrate and cleavage site; the star denotes the position of the 5′ label.
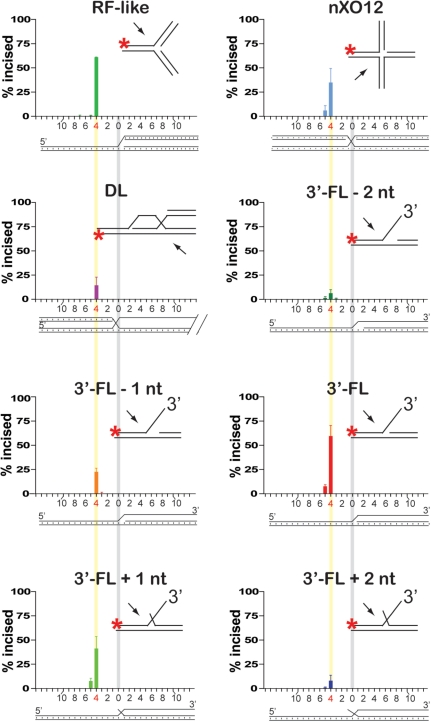
Figure 4.
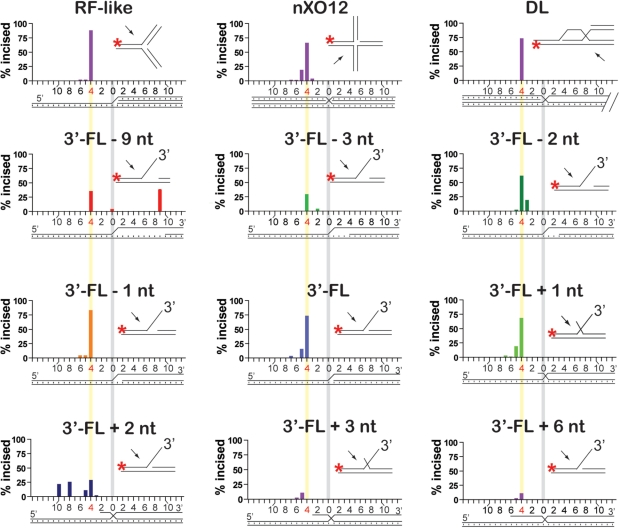
Mus81-Mms4 incises joint molecules at the branch point, adjacent to a phosphodiester backbone discontinuity. Quantitation of incision sites in Figure 3. Substrate branch point position is indicated by a vertical gray bar in all substrate schematics; the phosphodiester bond between the fourth and fifth nucleotides 5′ to the branch point is indicated by a vertical yellow bar.
Figure 5.
Mapping of Mus81-Mms4 incision sites at high protein to substrate ratio. Denaturing urea–PAGE analysis of substrate incision sites on RF-like, nXO12, D-loop (DL) and five 3′-FL-related structures. Assays were performed with 50 nM His10-FLAG-Mus81/GST-Mms4, 50 nM substrate, 30 min at 30°C. Where indicated, reactions were then supplemented with 0.5 mM ATP/3 mM Mg(OAc)2 and 10 U T4 DNA ligase, with incubation at room temperature for 15 min. All assays were terminated by boiling at 95°C, 2 min, with immediate transfer to ice. ‘L’ represents an oligonucleotide size ladder. The scheme on the right side of the gel illustrates the substrate and cleavage site; the star denotes the position of the 5′ label.
Figure 6.
Mus81-Mms4 incises joint molecules at the branch point, adjacent to a phosphodiester backbone discontinuity. Quantitation of incision sites in Figure 5. Substrate branch point position is indicated by a vertical gray bar in all substrate schematics; the phosphodiester bond between the fourth and fifth nucleotides 5′ to the branch point is indicated by a vertical yellow bar.
Joint molecule preparation
Substrate preparation was described by Ehmsen and Heyer (13), except in the case of incision site-mapping assays in which oligonucleotides upstream of the substrate branch point were 5′-phosphorylated: 250 pmol oligonucleotide was incubated with T4 PNK (New England Biolabs) in T4 DNA ligase buffer (containing 1 mM ATP), in a 50 µl volume for 30 min at 37°C followed by 10-min incubation at 65°C. Oligonucleotides were recovered using the Qiaquick® Nucleotide Removal Kit protocol (Qiagen). Complete phosphorylation was verified by denaturing urea–PAGE, which confirmed a greater electrophoretic mobility after addition of the negatively charged phosphate group. Substrates were annealed and purified as previously described, except for the modification that radiolabeled substrates were annealed with 50 pmol 50-mer and 100 pmol 25-mer, and nonradiolabeled substrates were annealed with 100–150 pmol 25-mer and 75 pmol 50-mer. Oligonucleotides used in substrate preparation were ordered without 5′-phosphate modification from Qiagen Operon. Nucleotide sequences are available on request (13).
Nuclease assays
Assays were performed as described by Ehmsen and Heyer (13). Heterodimer was diluted in standard enzyme diluent (SED: 10 mM Tris–HCl, pH 7.5, 0.5 mg/ml BSA) to an appropriate 10× reaction stock. Reactions were performed in buffer containing 25 mM HEPES pH 7.5, 3 mM MgOAc2, 1 mM DTT and 0.1 μg/ml BSA. For protein titrations on 3′-flap variants, three independent trials were performed with substrate concentration fixed at 50 nM and Mus81-Mms4 added to 5, 10, 20, 50 and 100 nM heterodimer. Reactions without protein were supplemented with standard enzyme diluent (SED: 10 mM Tris–HCl, pH7.5, 0.5 mg with pH7.5, 0.5 mg BSA). Reactions were pre-incubated in a 30°C water bath for 10 min, and were initiated by addition of an appropriate heterodimer dilution in SED with gentle mixing, followed by a 30-min incubation. Reactions were quenched by addition of 2 μl nuclease stop mix (200 mM EDTA, 2.5% SDS, 10 mg/ml proteinase K). Reactions were normalized for radioactivity and processed for electrophoresis as described for kinetic assays below. For kinetic analysis, at least three independent trials were performed for the substrates indicated in Figure 2 at 0.5, 1, 2.5, 5, 10, 20, 50 and 100 nM substrate and enzyme fixed at 5 nM, with all substrate concentrations processed in parallel. Reactions were preincubated in a 30°C water bath for 10 min, and were initiated by addition of 1 µl 50 nM heterodimer with gentle mixing. 0.5 µl aliquots were removed from each [substrate] reaction at 3, 6, 10, 15, 20, 30, 45 and 60 min and quenched immediately into pre-aliquoted 0.5 µl volumes of 0.5× nuclease stop mix. At each time point, 9 µl DNA loading dye (5% glycerol/bromophenol blue) were added and samples were electrophoresed by 10 × 20 cm native 10% TBE–PAGE at 100 V for 65 min. Gels were equilibrated in 5% glycerol or 3% glycerol/20% methanol for 30 min and vacuum-dried to Whatman® paper at 65°C, then exposed overnight to a phosphorimager screen. Reaction progress was quantified by Storm Phosphorimagery and ImageQuant software. Initial velocities were extrapolated from nonlinear regression curves defined by Graphpad Prism at 30 s reaction time. Michaelis–Menten analysis was performed using GraphPad Prism version 4.03 for Windows (GraphPad Software, San Diego, CA, USA; www.graphpad.com). Rates are expressed as nanomoles joint molecule substrate incised/minute, and where calculated for kcat, rates are expressed as number of joint molecule substrates incised per heterodimer molecule per minute.
Incision site determination
Nuclease assays were performed as previously described, with the exception that reaction volumes were 20 µl, at equimolar heterodimer: substrate (50 nM:50 nM) or at limiting heterodimer:substrate (5 or 10 nM:50 nM). Reactions were incubated for 30 min at 30°C. For assays followed by incubation with T4 DNA ligase, 9-ul reaction volume was transferred to a new 500 ul Eppendorf tube and 0.5 µl containing 60 mM Mg(OAc)2 and 10 mM ATP plus 0.5 µL (200 U) T4 DNA ligase (New England Biolabs) were added followed by incubation at room temperature for 15 min. For nicked duplex ligation controls, 50 nM nicked duplex (with or without 5′ phosphate at the internal nick) was incubated with T4 DNA ligase for 15 min, at room temperature. All reactions were stopped by denaturation at 95°C for 2 min, followed by transfer to ice. All samples were normalized for specific activity and loaded in 2 µl volumes onto 12% or 14% acrylamide/8 M urea denaturing PAGE. An oligonucleotide size ladder was run in parallel (olWDH775-784; sequences are available on request). Oligos were radiolabeled with γ32P-ATP, purified using a Qiaquick Nucleotide® Removal Kit or Microspin™ G-25 Sepharose columns (Amersham), and pooled to normalize specific activity after determination of activity by scintillation count. Incision site positions for the D-loop (DL) were extrapolated from a standard curve plotting ladder oligonucleotide sizes against migration distance.
RESULTS
Mus81-Mms4 prefers duplex DNA flush to the substrate branch point
Because FEN1/Rad27 is a 5′-flap endonuclease, one proposal for the synthetic lethality of the 5′-flap endonuclease with Mus81-Mms4/Eme1 in S. cerevisiae and S. pombe has suggested that the two nucleases function as alternative flap cutters (2,24–26). FEN1 is most active on a dually flapped structure, having a flap of 5′ polarity juxtaposed to a minor 3′ flap of single-nucleotide length (29,32). To test the model that Mus81 behaves as a 3′-flap endonuclease in a style similar to the 5′-flap endonuclease, we performed a kinetic analysis of Mus81-Mms4 activity on dually flapped substrates.
Unlike FEN1, Mus81-Mms4 activity is reduced on a dually flapped substrate (Figure 1). Of a panel of substrates related to the 3′-flapped joint molecule, the structure with duplex DNA flush to the branch point is processed most effectively. Mus81-Mms4 demands that the backbone discontinuity at the substrate branch point is unaltered by nucleotide additions to the deoxyribose at the 5′ position of the discontinuity that would overlap the 3′-flap. Mus81-Mms4 is more sensitive to extensions at the 5′ position of the nick than to retreat of the 5′ position from the branch point. Advancing duplex DNA from the branch point by 1–2 nt (which generates a short 5′ overhang) impairs substrate processing approximately ∼1.5–2.5-fold more than retreating duplex DNA from the branch point by 1–2 nt (which generates a small ssDNA gap) (Figure 1C). The overall DNA binding on these 3′-flap variants appears not considerably changed, as KM is not significantly altered when the duplex DNA is retreated or advanced by 1 nt from the branch point, and is nearly unaltered when the DNA is advanced or retreated by 2 nt (Figure 2). Instead, the processing impairment on these substrates is explained by a reduction in turnover. Altering the position of the duplex end by as little as one nucleotide reduces kcat, a reduction of 2.7-fold with a 1-nt flap, 5-fold with a 2-nt flap and nearly 25-fold with a 3-nt flap. Advancing the upstream duplex end as little as a single nucleotide of ssDNA reduces substrate processing by nearly 2-fold, and further advancing the upstream end into a 2-nt 5′ flap reduces substrate processing by up to 4-fold (Supplementary Table 1, Figure 2). We conclude that Mus81-Mms4 behaves very differently from FEN1 in its kinetic response to a dually flapped substrate. The -9-nt substrate (Figure 1A) showed evidence for an additional, novel cleavage site by Msu81-Mms4, which was mapped together with the cleavage sites of other substrates (see below; Figures 5 and 6).
Mus81-Mms4 substrate incision is determined by the position of the substrate branch point, not by the position of the upstream 5′ duplex end
To test the role of the upstream duplex in positioning Mus81-Mms4 incision, we determined the position of substrate incision on a number of DNA joint molecules. We incubated limiting heterodimer with excess substrate (10 nM heterodimer with 50 nM substrate; Figures 3 and 4) or equimolar heterodimer and substrate (50 nM heterodimer with 50 nM substrate; Figures 5 and 6) on nXO12 (nicked Holliday junction), RF-like (replication fork-like), DL (D-loop) and 3′-FL (3′-flap)-related structures, for 30 min at 30°C. Assays in the presence of limiting heterodimer were used to determine incision site positions under conditions that allow Mus81-Mms4 catalytic turnover; assays with stoichiometric quantities of heterodimer and substrate allowed the determination of incision site positions on an additional set of substrates that are too poorly incised for incision site quantitation under conditions of limiting heterodimer (specifically the 3′-flapped structures with 4-nt and 9-nt gaps, and 6-nt 5′-flap). We interpreted incision site positions by direct comparison of the incised oligonucleotide lengths to a radiolabeled ladder of oligonucleotide standards of known lengths (Figures 3 and 5). In all cases, incision takes place on the upper strand of the joint molecule (defined as the DNA backbone continuous with the 3′-flap in 3′-flapped structures), achieved by hydrolysis of the phosphodiester bond between the fourth and fifth nucleotides 5′ to the junction branch point (Figures 4 and 6). This is true whether the duplex DNA upstream of the branch point is advanced to a single-stranded 5′ DNA overhang or retreated to generate a small single-stranded DNA gap.
We tested whether the GST-tag on the Mms4 subunit influences cleavage efficiency or cleavage site selection but found no difference with heterodimer from which the GST-tag on Mms4 had been removed by PreScission protease treatment during the purification protocol (data not shown).
The position of incision is therefore determined strictly by the branch point, and not by the position of the upstream 5′ duplex end. Incision position is also not affected by the nature of duplex or single-stranded DNA arms that converge at the junction branch point, as all substrates (including the RF-like, D-loop and nicked Holliday junctions) are incised 4-nt 5′ of the branch point (Figures 3 and 4).
To further characterize the incision products of Mus81-Mms4, we incubated half of each incision reaction with T4 DNA ligase for 15 min at room temperature (25°C) and analyzed the products by gel electrophoresis in parallel. Incision that is not coincident with the junction branch point generates a small gapped region in the incision product, confirmed by the inability to ligate the resulting duplex with T4 DNA ligase. We demonstrated that a nicked duplex could be ligated by T4 DNA ligase under our reaction conditions, as 50 nM nicked duplex with a 5′-phosphate at the internal nick (prepared by oligonucleotide annealing) was readily ligated under identical circumstances, and this was dependent on the presence of the internal 5′-phosphate (nicked duplex prepared from nonphosphorylated oligonucleotides remained unligated; Figure 3). Ligatable nicked duplexes were not detected as products of Mus81-Mms4 incision on any of the substrates tested. We conclude that Mus81-Mms4 incision is determined by the branch point rather than by the position of the 5′-end (phosphorylated or unphosphorylated) at the junction.
To further establish the notion that Mus81-Mms4 cleavage depends on a branch point, we tested nicked duplexes, i.e. substrates lacking a branch point aside from a duplex discontinuity. Mus81-Mms4 very poorly cleaved nicked duplex substrates, as determined by native and denaturing analysis of products (data not shown).
DISCUSSION
Here we examined how S. cerevisiae Mus81-Mms4 responds to branch point properties of a DNA joint molecule substrate. We used kinetic parameters (KM, kcat) and incision site position to gauge the significance of an upstream duplex DNA end adjacent to the substrate branch point. These biochemical studies suggest an incision site determinant (the branch point position alone) that is an alternative to that which has been reported for Mus81-Mms4/Eme1 (the upstream duplex 5′-end) (17,25). Furthermore, we directly test the model that Mus81-Mms4 acts as a 3′-flapase, constituting a counterpart to the 5′-flapase activity of FEN1/Rad27 (2,24–26).
FEN1/Rad27 is a 5′-flap endonuclease; however, its preferred substrate is in fact dually flapped, having a single-nucleotide 3′-flap coincident with a longer 5′-flap (29,32). 5′-flapped substrates with a single-nucleotide 3′-flap are incised with up to 4-fold enhancement over unmodified 5′-flapped substrates (29), irrespective of the 3′-nt extension base identity (32). Whereas FEN1/Rad27 cleavage on a 5′-flapped structure with DNA flush to the branch point generates a mixed population of duplex products with incisions at several positions, incision on the dually flapped structure generates a homogeneous nicked duplex population that can be sealed by DNA ligase (29). Moreover, structural analysis of human FEN1 revealed a surface pocket that specifically interacts with the short but nontrivial 3′-flap, a DNA-binding site at the nuclease incision position that is conserved in archaeal FEN1 (33,34).
We show by kinetic analysis that Mus81-Mms4 acts very differently from FEN1/Rad27 on dually flapped structures, using variants of a 3′-flapped structure to test the consequence of advancing or retreating duplex end position at the branch point (Figures 1 and 2; Supplementary Table 1). Although the KM is relatively robust when the upstream duplex end position is altered by 1–2 nt, the reduced turnover (kcat) on these modified joint molecules indicates that Mus81-Mms4 is sensitive to even single nucleotide deviations in duplex DNA proximity to the substrate branch point (Figure 2). Substrate turnover (kcat) is reduced on these substrates when the duplex end position is altered by 1–2 nt (generating a small gap upstream of the branch point when retreated or a short 5′-flap when advanced). When the duplex end position is altered by more than 2 nt, substrate turnover drops dramatically and substrate binding becomes impaired. These results are consistent with the previous analysis by Bastin-Shanower et al. (25) showing that flaps with a flush end are the preferred Mus81-Mms4 substrate. Also, unlike FEN1/Rad27 on its preferred substrate, Mus81-Mms4 delivers incisions that are not directly ligatable (Figures 3–6). In addition, the recently solved crystal structure of a truncated hybrid zebra fish–human Mus81-Mms4 complex does not provide evidence for a pocket that could accommodate a dual flap like FEN1 (10). Another biochemical difference between Mus81-Mms4 and FEN1/Rad27 is the way the nuclease approaches the substrate. While FEN1/Rad27 approaches the junction from the single-stranded flap (30), Mus81-Mms4 approaches the junction from the duplex arms (10). The biochemical and structural data appear inconsistent with the model that Mus81-Mms4 acts as the 3′-flapase counterpart of the 5′-flapase Fen/Rad27.
Nevertheless, the 3′-flapase model provides a potentially satisfying explanation for the synthetic lethality between mutants in MUS81 and RAD27, particularly in light of the avid biochemical activity of Mus81-Mms4 on 3′-flap substrates (2,5,24–26). 5′- and 3′-flaps that are homologous to the double-stranded sequence they adjoin can isomerize in principle. Such isoenergetic isomerization may impose a strict demand for a 3′ or 5′-flapase, and cells could tolerate absence of one flapase but not of both. Five primary hypotheses have emerged to explain how 3′-flapped substrates may arise in vivo as structures potentially targeted by Mus81-Mms4, only two of which directly postulate that the 3′-flaps in question arise as a consequence of Rad51-mediated recombination: (i) flaps can be generated by strand displacement synthesis during DNA replication, known to be especially relevant during the removal of RNA primers that occur every 100–150 bp on DNA newly synthesized as lagging strands (30); (ii) flaps can be generated during removal of topoisomerase–DNA cleavage complexes, such as those irreversibly trapped by camptothecin-like inhibitors (35,36); (iii) flaps can be generated during Rad51-mediated DNA strand exchange (37,38); (iv) flaps can be generated by over-replication of DNA during any form of synthesis-dependent strand annealing (SDSA) (2,24,25); and (v) 3′-flaps occur during SSA, a Rad52-mediated reaction (37,38). While the contribution to flap cleavage by Mus81-Mms4 is unknown in some of these contexts, it has been shown that Mus81-Mms4 makes no contribution or a minimal contribution to the turnover of trapped topoisomerase–DNA cleavage complexes (35). In the context of Rad51-mediated strand invasion, either the exonuclease activity of DNA polymerase δ or Rad1-Rad10, the yeast XPF-ERCC1 paralog, cleaves a nonhomologous flap (39,40). Likewise, in the context of single-strand annealing (SSA), Rad1-Rad10 is needed for flap removal but not Mus81-Mms4 (35). Rather than signifying a role for Mus81-Mms4 as an alternative flap cutter in vivo, the synthetic lethality of the mus81 rad27 double mutant can also be interpreted in a different fashion. Mutations in the core recombination factors RAD51, RAD52, RAD55, RAD57 and RAD54 are all synthetically lethal with rad27 (41). Problems in Okazaki fragment processing in the absence of Rad27 may lead to lagging strand substrates such as single-stranded DNA flaps and gaps that require processing by recombination. This may be a context especially relevant for Mus81 function, as it appears likely that Mus81-Mms4 plays a specific role in homologous recombination at such replication-associated gaps (42). This interpretation is consistent with the observation that the synthetic lethality of mus81 sgs1 double mutants is suppressed by a defect in recombination (rad51, rad52, rad55, rad57 and rad54), suggesting that Mus81-Mms4 functions late in recombination in a pathway parallel to one controlled by Sgs1 (42). On the other hand, several lines of evidence suggest a function of Mus81-Mms4 in addition to the role downstream of Rad51 function, possibly in the cleavage of stalled replication forks to initiate recombination (43–45). In sum, although the available data cannot exclude a 3′-flapase function of Mus81-Mms4, there are no compelling genetic data to support this model. Structural observations (10) and now the biochemical argument presented here are inconsistent with Mus81-Mms4 being a 3′-flapase counterpart of the FEN1/Rad27 5′-flapase.
In addition to drawing a stark contrast to FEN1/Rad27 behavior on dually flapped substrates, the data presented here suggest that the branch point adjacent to a DNA-backbone interruption is a necessary and sufficient gauge from which Mus81-Mms4 phosphodiester bond hydrolysis is targeted 4 bp 5′ to the branch point. In a previous study mapping Mus81-Mms4 incision sites on 3′-flaps and other substrates, Bastin-Shanower et al. (25) suggested that the 5′ duplex end adjacent to the junction branch point directs the position of incision. Specifically, retreat of the duplex 5′ end upstream of the 3′-flap branch point was observed to result in a corresponding retreat in the position of incision by Mus81-Mms4, marked by a constant 5-nt distance between the 5′ duplex end and the position of branch point incision. The constant distance between upstream 5′ end and incision site was maintained until the branch point itself was reached, at which point further retreat of the 5′ end resulted in loss of incision on the substrate. In contrast, we found that regardless of the 5′ end distance from the branch point, nucleolytic incision is targeted primarily between the fourth and fifth phosphodiester bonds, 4-nt 5′ of the joint molecule branch point. This is true whether Mus81-Mms4 is at limiting or stoichiometric concentrations to joint molecule substrates (Figures 3–6), and whether or not the 5′ duplex end is phosphorylated (Figures 3–6, data not shown). Furthermore, this is true for heterodimer preparations purified by an approach that removes the GST tag on Mms4 (data not shown). The incision position therefore seems to be determined strictly by the branch point, and not by the position of the 5′ duplex end. Even when Mus81-Mms4 concentration is equimolar to substrate concentration and we could map incision positions on substrates with the upstream duplex retreated by up to 9 nt, most incisions take place 4-nt 5′ of the junction branch point (Figures 5 and 6). Furthermore, nicked duplex substrates that lack a branch point are nearly uncleaved by our preparation of S. cerevisiae Mus81-Mms4. The poor recognition and turnover of a nicked duplex underscores that a branch point is key to substrate processing (data not shown). Specifically, a DNA strand interruption (nick) flanked by upstream and downstream duplexes is not sufficient for incision. The reasons for this difference between our study and the previous study (25) are unclear. Sequence–context effects of the substrate can be excluded, as the same substrates were used in both studies. Different enzyme preparations (tags, purification host) were used, as well as different reaction conditions. Of particular importance is the enzyme to substrate ratio, which was low in our study due to the high specific activity of the enzyme preparations, which showed catalytic turnover, and much higher in the previous study (25). This may also explain that little incision of nicked substrates was observed in our study, whereas Bastin-Shanower et al. (25) observed cleavage of nicked duplexes by Mus81-Mms4. The same considerations can account for the difference to the study by Osman et al. (17), who reported that incision sites on nicked Holliday junction substrates responded to the position of the 5′ end.
Our incision site mapping, coupled with kinetic analysis on the same substrates, suggests a model for Mus81-Mms4 substrate processing that assigns kinetic importance to the upstream duplex as previously proposed, but relieves the duplex end position of a role in defining incision site location (Supplementary Figure 1). The nature of the flap (ssDNA in the 3′-flap or dsDNA in the RF-like substrate) does not influence cleavage kinetics or incision site selection [this study, (13)]. We observe that S. cerevisiae Mus81-Mms4 references the substrate branch point for positioning of its incision site, and depends on upstream duplex DNA flush to that branch point for most effective turnover. Our incision site mapping is consistent with an X-ray crystallographic structure of a partial vertebrate Mus81-Eme1 heterodimer (10). A DNA-binding surface on the nuclease domain presents a small knob that appears to anchor the substrate branch point and to splay duplex DNA downstream of the substrate branch point into a short stretch of ssDNA. The nuclease catalytic residues are precisely 4-nt 5′ of the anchored branch point. Incision several nucleotides 5′ of the branch point by Mus81-Mms4 has the consequence that the duplex products of its potential substrates (whether 3′-flap, nicked Holliday junction, replication fork, or D-loop in vitro) cannot be directly ligated—the 5′ end upstream of the branch point cannot be directly ligated to the 3′-hydroxyl product of Mus81-Mms4 incision. Whether this property is of biological relevance to the in vivo role of Mus81-Mms4 remains to be understood, and it is possible that the position of incision on substrates will differ in vivo in the context of other factors. Alternatively, substrate incision 4 nt from the branch point may be biologically important to minimize inappropriate ligation of duplex arms. Where multiple arms converge at a joint molecule branch point, especially when some of those arms remain in proximity after incision, off-target ligation may be avoided if incision generates a small gap between duplex arm pairs that should not be joined.
In summary, we report a kinetic analysis of S. cerevisiae Mus81-Mms4 on 3′-flap variants, revealing that the branch point is key to direct cleavage of joint molecules. These kinetic observations suggest that the mechanism by which Mus81-Mms4 processes DNA joint molecule substrates entails a sampling of branch point properties unlike those queried by FEN1/Rad27. Although both endonucleases recognize a structural branch point, FEN1/Rad27 targets a dually flapped substrate that occurs in the context of its biological function, whereas a dually flapped substrate impairs substrate processing by Mus81-Mms4. If Mus81-Mms4 targets 3′-flaps under any circumstances in vivo, it does so in a manner very different from FEN1/Rad27.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health Molecular & Cellular Biology training (T32 GM007377 to K.T.E.); and National Institutes of Health (GM58015 to W.-D.H). Funding for open access charge: National Institutes of Health (GM58015).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stephen Brill, Shannon Ceballos, Clare Fasching, Ryan Janke, Erin Schwartz, William Wright and Xiao-Ping Zhang for comments on the manuscript, and all members of the Heyer laboratory for support and discussion. William Wright is gratefully acknowledged for purification of Mus81-Mms4 without the GST tag. Jessica Sneeden is thanked for sequencing advice. We thank the Stephen Kowalczykowski, Carol Erickson and John Scholey laboratories for sharing instruments. K.T.E. specially thanks Toby Ehmsen.
REFERENCES
- 1.Ehmsen KT, Heyer WD. In: Recombination and Meiosis. Richard Egel DL, editor. Berlin-Heidelberg: Springer; 2008. pp. 91–164. [Google Scholar]
- 2.Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair. 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 4.Heyer WD, Ehmsen KT, Solinger JA. Holliday junctions in the eukaryotic nucleus: Resolution in sight? Trends Biochem. Sci. 2003;10:548–557. doi: 10.1016/j.tibs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 6.White MF. Archaeal DNA repair: paradigms and puzzles. Biochem. Soc. Trans. 2003;31:690–693. doi: 10.1042/bst0310690.. [DOI] [PubMed] [Google Scholar]
- 7.Ciccia A, Ling C, Coulthard R, Yan ZJ, Xue YT, Meetei AR, Laghmani EH, Joenje H, McDonald N, de Winter JP, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 9.Newman M, Murray-Rust J, Lally J, Rudolf J, Fadden A, Knowles PP, White MF, McDonald NQ. Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition. 2005;24:895–905. doi: 10.1038/sj.emboj.7600581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang JH, Kim JJ, Choi JM, Lee JH, Cho YJ. Crystal structure of the Mus81-Eme1 complex. Genes Dev. 2008;22:1093–1106. doi: 10.1101/gad.1618708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciccia A, Constantinou A, West SC. Identification and characterization of the human Mus81/Eme1 endonuclease. J. Biol. Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- 12.Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 13.Ehmsen KT, Heyer WD. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic structure-selective endonuclease. Nucleic Acids Res. 2008;36:2182–2195. doi: 10.1093/nar/gkm1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaillard P-H, Noguchi E, Shanahan P, Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- 15.Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogrunc M, Sancar A. Identification and characterization of human MUS81-MMS4 structure-specific endonuclease. J. Biol. Chem. 2003;278:21715–21720. doi: 10.1074/jbc.M302484200. [DOI] [PubMed] [Google Scholar]
- 17.Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role of Mus81-Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 18.Whitby MC, Osman F, Dixon J. Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 2003;278:6928–6935. doi: 10.1074/jbc.M210006200. [DOI] [PubMed] [Google Scholar]
- 19.Boddy MN, Gaillard P-HL, McDonald WH, Shanahan P, Yates JR, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 20.Chen X-B, Melchionna R, Denis C-M, Gaillard P-HL, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 21.Gaskell LJ, Osman F, Gilbert RJ, Whitby MC. Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates? EMBO J. 2007;26:1891–1901. doi: 10.1038/sj.emboj.7601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cote AG, Lewis SM. Mus81-dependent double-strand DNA breaks at in vivo-generated cruciform structures in S. cerevisiae. Mol. Cell. 2008;31:800–812. doi: 10.1016/j.molcel.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 24.de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de los Santos T, Loidl J, Larkin B, Hollingsworth N. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics. 2001;159:1511–1525. doi: 10.1093/genetics/159.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fricke WM, Bastin-Shanower SA, Brill SJ. Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair. 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Tong AHY, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CWV, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2363–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 29.Kao KI, Henricksen LA, Liu Y, Bambara RA. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 31.Xie Y, Liu Y, Argueso JL, Henricksen LA, Kao HI, Bambara RA, Alani E. Identification of rad27 mutations that confer differential defects in mutation avoidance, repeat tract instability, and flap cleavage. Mol. Cell Biol. 2001;21:4889–4899. doi: 10.1128/MCB.21.15.4889-4899.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaiser MW, Lyamicheva N, Ma W, Miller C, Neri B, Fors L, Lyamichev VI. A comparison of eubacterial and archaeal structure-specific 5′-exonucleases. J. Biol. Chem. 1999;274:21387–21394. doi: 10.1074/jbc.274.30.21387. [DOI] [PubMed] [Google Scholar]
- 33.Toueille M, El-Andaloussi N, Frouin I, Freire R, Funk D, Shevelev I, Friedrich-Heineken E, Villani G, Hottiger MO, Hubscher U. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapados BR, Hosfield DJ, Han S, Qiu JZ, Yelent B, Shen BH, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 35.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl Acad. Sci. USA. 2002;99:13669–13674. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connelly JC, Leach DRF. Repair of DNA covalently linked to protein. Mol. Cell. 2004;13:307–316. doi: 10.1016/s1097-2765(04)00056-5. [DOI] [PubMed] [Google Scholar]
- 37.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heyer WD. In: Molecular Genetics of Recombination. Aguilera A, Rothstein R, editors. Berlin-Heidelberg: Springer; 2007. pp. 95–133. [Google Scholar]
- 39.Ivanov EL, Haber JE. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Symington LS. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 1998;26:5589–5595. doi: 10.1093/nar/26.24.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ii M, Brill SJ. Roles of SGS1, MUS81, and RAD51 in the repair of lagging-strand replication defects in Saccharomyces cerevisiae. Curr. Genet. 2005;48:213–225. doi: 10.1007/s00294-005-0014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Froget B, Blaisonneau J, Lambert S, Baldacci G. Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint. Mol. Biol. Cell. 2008;19:445–456. doi: 10.1091/mbc.E07-07-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.