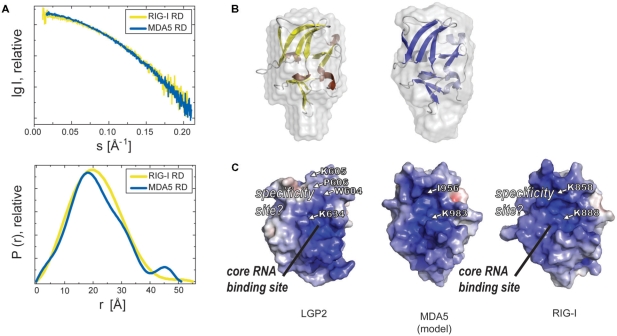
Figure 5.
Comparison of RDs of RIG-I-like helicases. (A) Small angle X-ray scattering curves of RIG-I RD and MDA5 RD (upper panel, merged from curves obtained with protein concentrations of 2, 5 and 10 mg/ml). The lower panel shows the pair distribution functions for RIG-I RD and MDA5 RD derived from raw data with Gnome (ATSAS 2.1). (B) Superimposition of SAXS surface models of RIG-I (left) and MDA5 RD (right) with crystal structures of RIG-I RD (yellow, brown) and a model of MDA5 RD (model based on RIG-I and LGP2 RD crystal structure, blue, light blue), respectively. (C) Electrostatic surface potential (ranging from blue=9 kT/e to red=−9 kT/e) of LGP2 RD, RIG-I RD (from crystal structure) and a model of MDA5 RD. Residues crucial for general RNA interaction and either 5′-triphosphate RNA specificity in RIG-I (K888, buried K858) or dsRNA specificity in LGP2 (corresponding K634, P606 and additionally buried W604 and K605) are highlighted as well as corresponding residues for MDA5 RD (K983, buried I956).