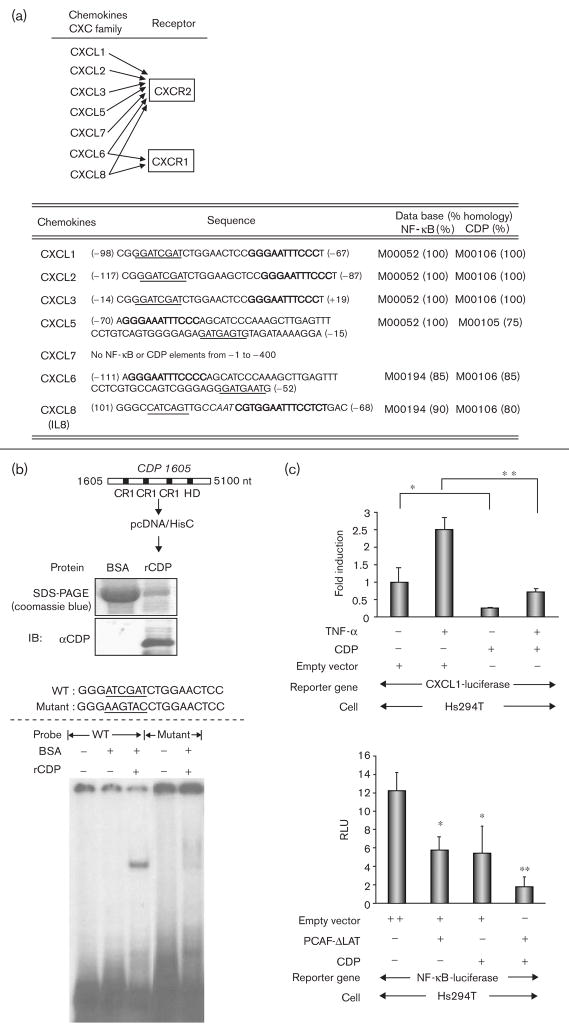
Fig. 2.
Putative CCAAT displacement protein (CDP) sites are near the nuclear factor-kappa beta (NF-κB) sites in the promoters of chemokines for CXCR1 and CXCR2. (a) Top: CXC ligands for CXCR1 and CXCR2. Bottom: the sequences of the putative NF-κB and CDP binding sites in the promoters of the CXC ligands. The promoter ID numbers and the percentages of homologies are followed by the TFSEARCH (www.cbrc.jp). Block letters: NF-κB binding sequences; underlined letters: CDP binding sequences; Italic letters: CCAAT sequence. (b) Top: purified DNA-binding domain of CDP. CDP cDNA (from + 1605 to + 5100, acc. no.: M74099) containing DNA binding domains (CR1-3 and homeodomain) was subcloned into the expression vector pcDNA/HisC. The expressed CDP recombinant protein (5 μg) was stained with coomassie blue after sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and subjected to immunoblot analysis with the αCDP antibody. CR, cut repeat; HD, homeodomain; nt, nucleotide. Bottom: electrophoretic mobility shift assay (EMSA) using a representative CDP binding sequence as indicated. The binding of the CDP recombinant protein was analyzed in EMSA using the CDP binding sequence from the CXCL1–3 promoters. (c) Top: cells were seeded in 24-well plates and the transfection of the reporter constructs were the same as stated in Fig. 1e. Cells were cotransfected with 0.2 μg/well of CDP expression plasmid or control empty plasmid. The following day, cells were incubated in serum-free medium (SFM) for 16 h and further incubated with/without TNF-α (10 ng/ml) for 4 h in SFM. The normalization of the transfection efficiency, and data analysis were the same as stated in Fig. 1e. The assays were done thrice. *P < 0.05; **P < 0.01 (Student’s t-test). Bottom: cells were cotransfected with following transactivators: (i) 0.2 μg/well of empty vector, (ii) 0.1 μg/well of empty vector + 0.1 μg/well of CDP expression plasmid, (iii) 0.1 μg/well of empty vector + 0.1 μg/well of PCAF-ΔLAT, and (iv) 0.1 μg/well of CDP expression plasmid + 0.1 μg/well of PCAF-ΔLAT. The measurement of luciferase activities and the data analysis are the same as above. *P < 0.05; **P < 0.01 (Student’s t-test).