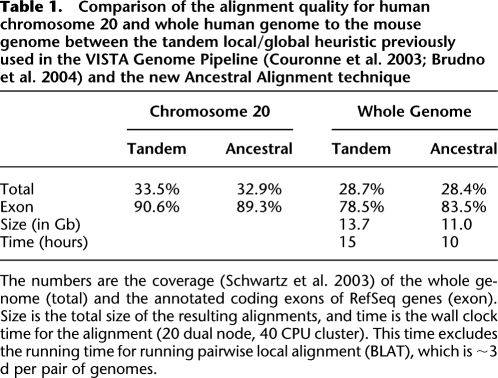
Table 1.
Comparison of the alignment quality for human chromosome 20 and whole human genome to the mouse genome between the tandem local/global heuristic previously used in the VISTA Genome Pipeline (Couronne et al. 2003; Brudno et al. 2004) and the new Ancestral Alignment technique
The numbers are the coverage (Schwartz et al. 2003) of the whole genome (total) and the annotated coding exons of RefSeq genes (exon). Size is the total size of the resulting alignments, and time is the wall clock time for the alignment (20 dual node, 40 CPU cluster). This time excludes the running time for running pairwise local alignment (BLAT), which is ∼3 d per pair of genomes.