Figure 6.
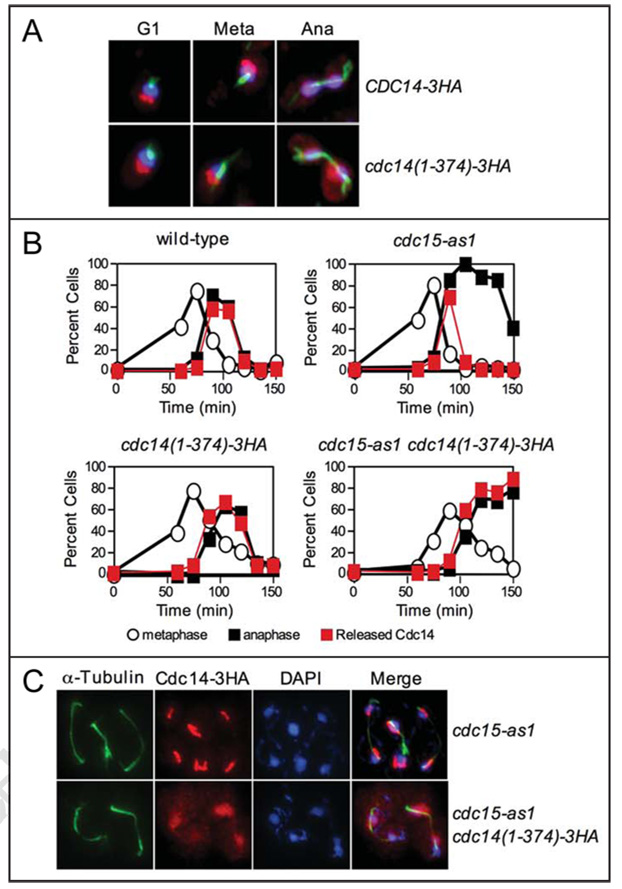
The C-terminus of Cdc14 is required for proper localization of Cdc14 during anaphase. (A) The subcellular localization of Cdc14-3HA (A1411) and Cdc14(1–374)-3HA (A19448) fusions in exponentially growing cells. Cdc14 is shown in red, microtubules are shown in green, and DNA is shown in blue. (B) mad1Δ cells carrying a Cdc14-3HA fusion (A2853), mad1Δ cdc15-as1 cells carrying a Cdc14-3HA fusion (A21212), mad1Δ cdc14(1–374)-3HA cells (A20792), and mad1Δ cdc15-as1 cdc14(1–374)-3HA cells (A21215) were grown and arrested in G1 as described in Figure 1B. MAD1 was deleted in these strains to ensure that the spindle assembly checkpoint did not interfere with cell cycle progression. After 2.5 hours, cells were washed with 10 volumes YEP and released into pheromone-free YEPD media containing 10 µM cdc15-as1 inhibitor (PP1 analog 8). After 100 minutes, alpha factor (10 µg/ml) was re-added to prevent cells from entering a new cell cycle. The percentage of cells with metaphase spindles (open circles), anaphase spindles (closed black squares), and Cdc14 released from the nucleolus (closed red squares) was determined at the indicated times. At least 100 cells were counted at each time point. (C) The subcellular localization of Cdc14 in mad1Δ cdc15-as1 carrying a Cdc14-3HA fusion and mad1Δ cdc15-as1 cdc14(1–374)-3HA cells 135 min after release from a G1 arrest. Cdc14 is shown in red, microtubules are shown in green, and DNA is shown in blue.