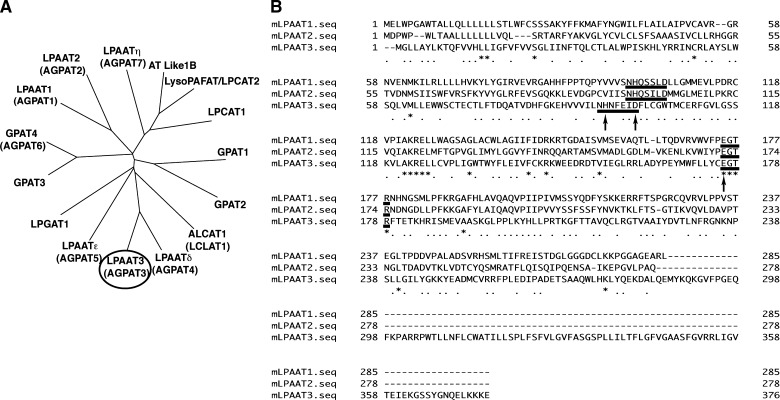
Fig. 1.
Phylogenetic tree of AGPAT family members and alignment of mLPAAT1, 2, and 3. A: A phylogenetic tree was drawn by using ClustalW, DDBJ (http://www.clustalw.ddbj.nig.ac.jp/top-j.html). Sequences of mouse acyltransferases are available in the DDBJ/EMBL/GenBank databases. mLPAAT3 is circled. B: mLPAAT1, mLPAAT2, and mLPAAT3 sequences were aligned using Genetic-Mac software. Conserved putative catalytic motif NHX4D and binding motif EGTR are underlined. Mutated amino acids are indicated by arrows (see Fig. 5). The accession numbers are shown as follows: GPAT1 (NP_032175), GPAT2 (NP_001074558), GPAT3 (NP_766303), GPAT4 (NP_061213), LPAAT1 (NP_061350), LPAAT2 (NP_080488), LPAAT3 (AB377215), LPGAT1 (NP_758470), ALCAT1 (acyl-CoA:lysocardiolipin acyltransferase 1; also called as LCLAT1) (Q3UN02), LPCAT1 (BAE94687), LysoPAFAT/LPCAT2 (BAF47695), LPAATδ (NP_080920), LPAATɛ (NP_081068), LPAATη (NP_997089), and AT Like 1B (NP_081875).