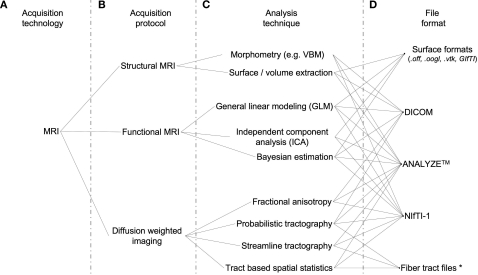
Figure 2.
Data handling complexity in MRI analysis streams. A schematic representation of the some of the levels of abstraction considered when preparing software capable of handling multi-modal neuroimaging data. (A) The technology type used: Here we use MRI as an example. (B) Some MRI acquisition protocols or sub-types: a researcher using a combination of protocols may, for example, be looking for changes in blood oxygenation using functional MRI, localizing the regions of activation to specific brain regions using structural MRI, and then looking for anatomical connections between these regions using Diffusion weighted MRI. They may then wish to overlay the results from each modality to explore spatial relationships. (C) Examples of the types of different analysis algorithms and routines for any given protocol. (D) Examples of data formats: although researchers may use the same technology, the same protocol, and even the same analysis technique/algorithm, they may save their results in different file formats not immediately accessible to software utilized at other sites. *In the case of Fiber tract files, few standard file formats have been developed specifically for DTI data, and even fewer for saving the results of fiber tracking algorithm output. The.nrrd file format (http://www.na-mic.org/Wiki/index.php/NAMIC_Wiki:DTI:Nrrd_format) is used by 3D Slicer to load DTI values and parameters into memory. Fibers are subsequently calculated and can be saved to a vtk file format, unspecific for DTI fibers but useful for import and conversion by any VTK based programs, including DV3D.