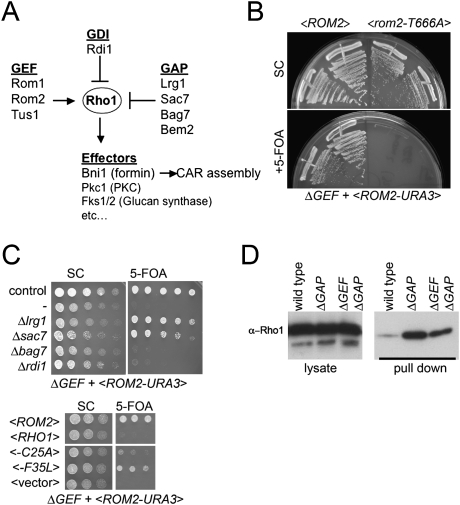
Figure 2.
Creation of viable yeast strains lacking all Rho1 GEFs. (A) Outline of Rho1 regulation in budding yeast. See the text for details. (B) A plasmid shuffle assay demonstrates that the essential function of Rho1 GEFs requires their catalytic activity. The ΔGEF strain grows in the presence of a <ROM2 URA3 CEN> plasmid but fails to grow after counterselection with 5-FOA. A point mutation in a critical catalytic residue in the DH domain of Rom2 (T666A) fails to complement the lethality of the ΔGEF strain. The indicated strains were grown for 3 d at 24°C. (C) GAP gene deletions or rapid cycling Rho1 mutants restore viability to the ΔGEF strain. A plasmid shuffle was performed to assess viability as in B. (Top) Deletion of the Rho1 GAP genes LRG1 or SAC7 (but not BAG7) restored growth to the ΔGEF strain at 24°C. Deletion of the sole Rho1 GDI gene RDI1 had no effect on growth of the ΔGEF strain. (Bottom) Two predicted Rho1 “rapid cycling” mutations restore growth to the ΔGEF strain. (D) The ΔGEF ΔGAP strain has high levels of Rho1-GTP; deletion of Rho1 GEFs has only a subtle effect on the total cellular levels of Rho1-GTP. GTP-Rho1 was assayed in the indicated strains by a Rho-binding domain pull-down assay.