Abstract
Tissue factor pathway inhibitor-2 (TFPI-2) is a serine proteinase inhibitor that induces caspase-mediated apoptosis when offered to a variety of tumor cells. In order to investigate the mechanism of TFPI-2-induced apoptosis, we initially studied the uptake and trafficking of TFPI-2 by HT-1080 cells. Exogenously offered TFPI-2 was rapidly internalized and distributed in both the cytosolic and nuclear fractions. Nuclear localization of TFPI-2 was also detected in a variety of endothelial cells constitutively expressing TFPI-2. Nuclear localization of TFPI-2 required a NLS sequence located in its Lys/Arg-rich C-terminal tail comprising residues 212-233, as a TFPI-2 construct lacking the C-terminal tail failed to localize to the nucleus. Complexes of TFPI-2 and importin-α were co-immunoprecipitated from cell lysates of HT-1080 cells either offered or overexpressing this protein, providing evidence that TFPI-2 was shuttled to the nucleus by the importin system. Our results provide the initial description of TFPI-2 internalization and translocation to the nucleus in a number of cells.
Keywords: TFPI-2, serine proteinase inhibitor, extracellular matrix, nuclear localization, importins
Introduction
Tissue factor pathway inhibitor-2, a 32 kDa Kunitz-type serine proteinase inhibitor, is primarily synthesized and secreted into the extracellular matrix (ECM) by a wide variety of cells including keratinocytes [1], dermal fibroblasts [1], endothelial cells [2], smooth muscle cells [3] and synoviocytes [4]. Several lines of evidence suggest that TFPI-2 regulates the plasmin-mediated activation of matrix pro-metalloproteinases and plays a significant role in the regulation of ECM degradation, which is an essential step for tumor cell invasion and metastasis [5-7].
We have recently demonstrated that exogenous application of recombinant TFPI-2, as well as a mutated first Kunitz-type domain (R24K KD1), induced caspase-mediated apoptosis in several tumor cell lines through a mechanism that most likely involved their serine proteinase inhibitory activity [8]. In the course of these studies, we observed that a portion of the TFPI-2 offered to the HT-1080 fibrosarcoma cells was internalized, as the cell lysate contained significant amounts of immunoreactive TFPI-2 following a 48 h incubation [8]. The present study was initiated to further investigate the intracellular distribution of offered TFPI-2 in cell lines that do not secrete this protein, and compare these patterns with cells that either constitutively synthesize TFPI-2 or stably transfected to overexpress this protein. Using immunoblotting and immunocytochemistry approaches, we demonstrate that offered TFPI-2 is rapidly internalized in cells that do not constitutively synthesize this protein and is translocated to the nucleus. Nuclear localization of TFPI-2 was also observed in cells that constitutively synthesize TFPI-2, as well as cells overexpressing this molecule. We further report that the TFPI-2 C-terminal tail contains a putative bipartite nuclear localization signal (NLS), and that a truncated TFPI-2 preparation lacking this tail was not detected in the nucleus. Moreover, evidence is presented that the C-terminal tail of TFPI-2 also appears to play a role in the transduction of TFPI-2 through the cell membrane. While the functional role of TFPI-2 in the nucleus is unknown, our working hypothesis predicts that, like several other proteinase inhibitors previously shown to be nuclear-associated [9-13], TFPI-2 probably regulates one or more serine proteinases involved in proteolytic degradation of nuclear components.
Materials and Methods
Cell lines and reagents
The human fibrosarcoma cell line (HT-1080) and primary embryonal kidney cell line (HEK 293), were obtained from American Type Culture Collection (Manassas, VA). Human umbilical vein endothelial cells (HUVECs) were obtained from Cambrex (Walkersville, MD). Human aorta endothelial cells (HAEC) and dermal capillary endothelial cells (DMEC), were obtained from Cell Systems (Kirkland,WA). Dulbecco's minimal essential medium (DMEM), penicillin, streptomycin, protease inhibitor cocktail, murine anti-human importin-α antibody, and murine anti-human alpha-tubulin antibody were purchased from Sigma-Aldrich (St Louis, MO). Anti-Histone H1 antibody was obtained from Gene Tex (San Antonio, TX). Fetal bovine serum was obtained from Hyclone (Ogden, UT). Nitrocellulose (NC) membranes, goat anti-rabbit IgG-HRP, goat anti-mouse IgG-HRP, and AffiGel 10 were obtained from Bio-Rad (Hercules, CA). Alexa Fluor 555-conjugated goat anti-mouse IgG was generously provided by Dr. Bridget Wilson. Alexa Fluor 488 protein labeling kit was purchased from Invitrogen (Carlsbad, CA). Mounting media with DAPI was from Vector Laboratories (Burlingame, CA). Chemiluminescent HRP substrate was purchased from Millipore Corporation (Billerica, MA). Two-chamber culture slides were obtained from BD Bioscience (Bedford, MA). Profound™ co-immunoprecipitation kit was purchased from Pierce (Rockford, IL). All other reagents were of the highest quality commercially available.
Cell culture
HEK 293 and HT-1080 cell lines were maintained in Dulbecco's minimal essential medium (DMEM), supplemented with heat-inactivated 10% fetal bovine serum and penicillin-streptomycin. The cells were cultured at 37°C in a humidified atmosphere containing 6% CO2. Similarly, endothelial cells were maintained in medium 199 supplemented with 20% fetal bovine serum, 100 μg/ml heparin, 100 μg/ml ECGS and penicillin-streptomycin.
Construction of plasmids and recombinant protein preparation
A TFPI-2 construct lacking the C-terminal tail (TFPI-21-188) was generated by PCR amplification using pcDNA3-TFPI-2 as the template [6], and the resulting amplicon subcloned into the EcoRI site of the pcDNA3.0 expression vector. An R24K KD1-C-tail chimera (R24K KD1-CT) was prepared by ligating the KD1 and C-tail fragments generated by PCR amplification. The KD1 fragment was amplified from pET-R24K KD1 [14] using primer sets containing NdeI and EcoR I restriction sites, while the C-tail was derived from the pcDNA3-TFPI-2 [6] using a primer set containing EcoR1 and Xho I restriction sites. Following amplification, the PCR products were digested with EcoRI and ligated to generate R24K KD1-CT, which was subsequently inserted into the Nde I/ Xho I site of the pET28a expression vector.
HT-1080 and HEK 293 cell lines were stably transfected to overexpress either wild-type human TFPI-2 or a mutant TFPI-2 construct lacking the C-terminal tail (TFPI-2 1-188), and were maintained as described [6, 15]. An anti-human TFPI-2 murine monoclonal antibody designated as SK-9 was prepared as described [15] and coupled to Affi-Gel 10 according to the manufacturer's recommendation. Recombinant human TFPI-2 was purified from HEK 293 serum-free conditioned media by a two-step chromatography procedure involving heparin-agarose [17] and SK-9-AffiGel 10 affinity chromatography. In the latter procedure, heparin-agarose purified TFPI-2 was dialyzed against 50 mM Tris-HCl (pH 7.5) and applied to the SK-9-AffiGel 10 column equilibrated at room temperature with this buffer. After a wash step with 50 mM Tris-HCl (pH 7.5)/0.5 M NaCl, TFPI-2 was eluted with 0.1 M glycine (pH 2.5)/0.5 M NaCl into one-tenth volume of 1 M Tris-HCl (pH 8.8) to immediately neutralize the pH 2.5 glycine. Recombinant TFPI-21-188 was expressed in stably-transfected HEK 293 cells and purified from the HEK 293 serum-free conditioned media by a combination of SP-Sepharose chromatography and SK-9-Affi-Gel10 immunoaffinity chromatography as described above. Recombinant R24K KD1 and R24Q KD1 were prepared as previously described [14]. Recombinant R24K KD1-CT was expressed in E.coli and purified as described for R24K KD1 [14].
Treatments of HT-1080 cells with recombinant proteins
HT-1080 cells were grown in 6 well plates under standard conditions. At confluence, the cells were treated with fresh medium containing either wild-type TFPI-2, TFPI-21-188, R24Q KD1, R24K KD1, or R24K KD1-CT as previously described [8]. Briefly, duplicate wells were treated with purified proteins (1μM final concentration) and incubated at either 37°C or 4°C for different time periods. Two wells were also treated with PBS at each temperature to serve as a control. At selected time points, the media was removed and the cells were rinsed once with PBS. The cells were then washed with 1M NaCl/PBS for 30 minutes with gentle shaking to dissociate cell surface-bound proteins [2]. Finally, the cells were rinsed once with PBS, trypsinized and harvested for the preparation of cell lysates and cell fractions.
Preparation of cell lysates and cell fractions
To prepare total cell lysates, 1-3×106 cells were lysed by sonication in 500 μl of lysis buffer containing of 125mM Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 50 mM sodium phosphate, 1mM PMSF and protease inhibitor cocktail. The lysate was kept on ice for about 10 minutes, centrifuged for 15 min at 10,000×g at 4°C, and the supernatant recovered. To prepare nuclear and cytosolic fractions, ∼2 × 106 cells were harvested and washed twice with cold PBS by centrifugation at 600×g in a Beckman J-6M/E centrifuge for 7 min at 4 °C. Five volumes of ice cold cytosolic buffer (10mM Hepes pH 7.4, 0.33M sucrose, 1mM MgCl2, 0.1% Triton X-100 and protease inhibitor cocktail) was added to the cells and incubated on ice for 15 min. The cytosolic fraction was collected by centrifugation at 900×g for 5 min at 4°C. The resulting undissolved pellet was washed twice with cytosolic buffer followed by centrifugation at 900×g for 5 min at 4°C. Finally, the resulting pellet was resuspended in 5 volumes of ice cold buffer containing 0.45M NaCl in 10mM Hepes (pH 7.4) and protease inhibitor cocktail. The suspension was incubated on ice for an additional 15 min to dissolve the nucleus, and subsequently centrifuged at 18,000×g for 5 minutes at 4°C. The resulting supernatant was collected as the nuclear extract. All samples were boiled for 3 min in the presence of 5% SDS and stored at -20 °C until use.
Immunoblot analyses
The cell lysate, cytosol and nuclear fractions were subjected to SDS-PAGE using 4-20% polyacrylamide gradient gels. Following electrophoresis, the proteins were electrotransferred to nitrocellulose membranes and subsequently blocked with 5% blotting grade non-fat dry milk in TBS/0.1% Tween- 20 at room temperature for 2 h. The membranes were then probed with specific antibodies dissolved in fresh blocking buffer. Immunoreactive proteins were identified using HRP-conjugated secondary antibodies and a chemiluminescent reagent system essentially as described [18]. The integrity of the cytosolic and nuclear fractions was verified using anti-alpha-tubulin and anti-histone-H1 antibodies, respectively [19].
Confocal Microscopy
The internalization of recombinant TFPI-2 in HT-1080 cells was assessed by confocal microscopy using a Zeiss LSM510-META microscope and Alexa Fluor 488-conjugated TFPI-2. Approximately 0.5 mg of recombinant TFPI-2 protein was labeled with Alexa Fluor 488 dye following the manufacturer's instructions. Alexa Fluor-conjugated protein (50 μg/ml) was added to two sets of HT-1080 cells that were grown to confluence in two-chamber culture slides and incubated either at 37°C or 4°C for different time periods. Cells were then washed twice with cold PBS and fixed with 4% paraformaldehyde solution for 15 minutes. The slides were further rinsed twice with cold PBS, treated with a drop of mounting medium containing DAPI, and covered with a coverslip for confocal microscopy.
In order to view the cellular localization of endogenously produced TFPI-2 either in endothelial cells or cells stably-transfected with human TFPI-2 and TFPI-21-188 cDNA, cells were grown in two-chamber culture slides as described above. Cells were rinsed twice with PBS and fixed in 4% paraformaldehyde solution for 30 minutes at room temperature. Following three washes with PBS, cells were permeabilized in 0.1% Triton X-100 in PBS for 10 minutes and further rinsed three times with PBS. Cells were then blocked with 10% goat serum in PBS for 1 h at room temperature and incubated for 2 h at room temperature with 10 μg/ml of SK-9, a murine monoclonal antibody raised against TFPI-2 [16]. Cells were then washed with 1% goat serum in PBS three times for 10 min, and incubated with goat anti-mouse Alexa Fluor-555 (1:250) for an additional 2 h at room temperature in the dark. Finally, the cells were washed five times with 1% goat serum in PBS for 15 min and a drop of mounting medium containing DAPI was added and covered with coverslip in preparation for confocal microscopy.
Co-immunoprecipitation assay
To identify the adapter protein(s) involved in transporting TFPI-2 to the nucleus, we performed co-immunoprecipitation studies using the Profound™ co-immunoprecipitation kit and cell lysates derived from either vehicle-treated HT-1080 cells, TFPI-2-treated HT-1080 cells or HT-1080 cells overexpressing TFPI-2. Approximately 4.5 × 106 cells from each system were washed twice with PBS, and the lysates prepared in the presence of proteinase inhibitors using the mammalian protein extraction reagent provided in the kit. Complexes of importin-α and TFPI-2 were removed from the lysate by co-immunoprecipitation according to the manufacturer's instructions. In this procedure, antibodies specific for either importin-α or TFPI-2 (SK-9) were covalently linked at room temperature for 16 h to an amine-reactive gel in coupling buffer (8 mM sodium phosphate/2 mM potassium phosphate/10 mM KCl/140 mM NaCl [pH 7.4]). Cell lysates from the above preparations were added to the antibody-coupled columns and incubated at 4°C for 18 h with gently end-over-end mixing. The columns were then washed several times with coupling buffer containing 0.5 M NaCl to remove non-specifically bound proteins, and finally eluted with the elution buffer provided in the kit. The eluted immunoprecipitation complexes were neutralized with 1 M Tris-HCl (pH 8.8), treated with 0.2 volumes of the non-reduced SDS sample buffer (0.3 M Tris-HCl (pH 6.8)/5% SDS/50% glycerol), boiled for 5 min, and subjected to immunoblotting.
Immunoblot densitometry analysis
The relative intensity of the immunoblot bands were quantified using Quantiscan software (Biosoft™). The intensities are reported in arbitrary units (A.U.) obtained by subtracting the background value from the corresponding band's mean value.
Results
Internalization of exogenous TFPI-2 by HT-1080 fibrosarcoma cells
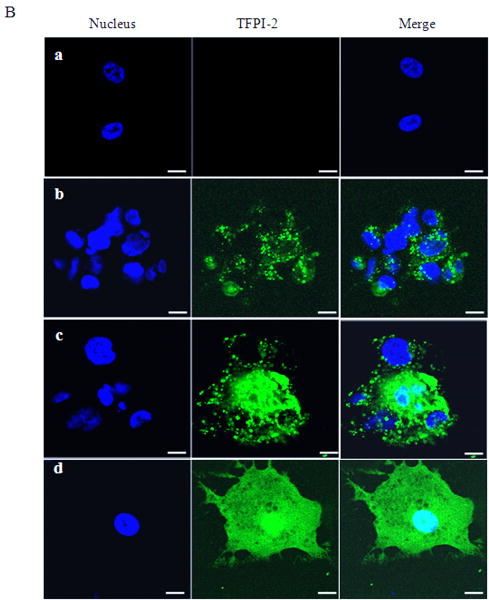
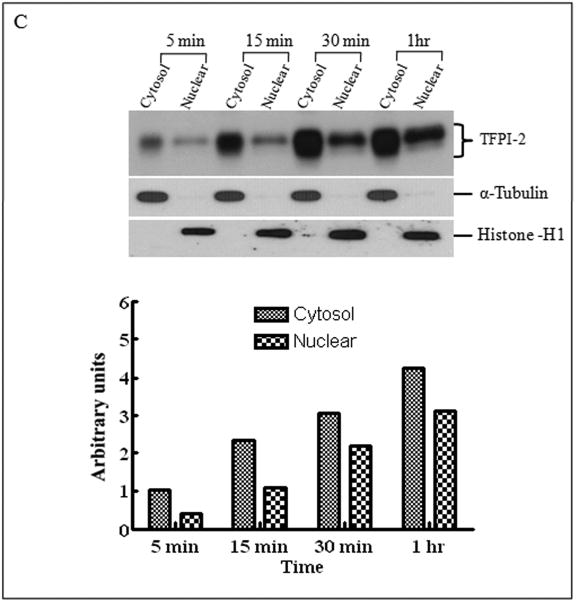
In a previous study, we reported that exogenous TFPI-2 induced caspase-mediate apoptosis in several human tumor cell lines [8]. Moreover, recombinant TFPI-2 offered to a human fibrosarcoma cell line, HT-1080, was internalized by these cells but was not degraded intracellularly [8]. To assess the functional significance of TFPI-2 internalization by these cells, we initially incubated HT-1080 cells with 1 μM TFPI-2 for various time periods (24-72 h) and subsequently assessed the intracellular distribution of the internalized TFPI-2. Using immunoblotting techniques, we observed that TFPI-2 was internalized and distributed in the cytosolic, as well as the nuclear fractions. Maximal cytosolic and nuclear localization of TFPI-2 by these cells occurred at 48 h, and diminished after 72 h of incubation (data not shown). To investigate the rate at which TFPI-2 was internalized and translocated to the nucleus, we offered TFPI-2 (1 μM) to the HT-1080 cells and temporally assessed TFPI-2 intracellular distribution within the first 15 min of offering. Figure 1A demonstrates that TFPI-2 is rapidly internalized by these cells and translocated to the nucleus within minutes of offering. The integrity of each cellular fraction was verified by separate immunoblotting experiments of these fractions using anti-α-tubulin IgG and anti-histone H1 IgG as cytosolic and nuclear markers, respectively (Fig. 1A). Consistent with the immunoblotting results, confocal microscopy, using Alexa Fluor 488-conjugated TFPI-2, revealed cytosolic and nuclear localization of TFPI-2 following a 2 h incubation with the HT-1080 cells at 37°C, whereas vehicle-treated cells revealed no fluorescence under identical conditions (Fig. 1B). Internalization and translocation of TFPI-2 to the nucleus was temperature-dependent, as the amount of TFPI-2 localized in the nucleus in 60 min at 4°C was roughly equivalent to that observed in 5 min at 37°C (Fig. 1C). In addition, confocal microscopy of Alexa Fluor 488-conjugated TFPI-2 incubated for 5 min, 10 min and 2 h at 4°C revealed a limited punctate distribution of TFPI-2 in the cytosol at 5 and 10 min. At the 2 h time point at 4°C, significant cytosolic and nuclear-associated TFPI-2 was also observed (Fig 1D). In contrast, uniform distribution of TFPI-2 was observed in cells at 37°C at the 2 h time point (Fig. 1B). In contrast to the complete TFPI-2 molecule, R24K KD1 and R24Q KD1, each at 1 μM concentrations, failed to be internalized by HT-1080 cells following a 48 h incubation at 37°C (data not shown). This result provided suggestive evidence that either the intact molecule or other TFPI-2 domains were required for cell binding and/or internalization.
Figure 1. Internalization of recombinant TFPI-2 protein in HT-1080 cells.
Cells were grown to confluence under standard conditions either in 6 well plates or two-chamber culture slides. (A), Cells grown in 6 well plates were treated with 1 μM TFPI-2 and incubated at 37°C for the indicated times and harvested for fractionation. To detect the presence of offered proteins, cytosolic and nuclear fractions were probed with anti-TFPI-2 antibody as described in Methods. The purity of cytosolic and nuclear fractions was verified using anti-alpha-tubulin and anti-histone-H1 antibodies, respectively. For immunoblot analysis, the intensity of blot bands were assessed by densitometric semi-quantitation and depicted by a bar diagram (lower panel). (B), Cells grown in two-chamber culture slides were incubated with either vehicle control or 50 μg/ml of Alexa Fluor-conjugated TFPI-2 at 37°C for the indicated times and processed for confocal microscopy. (a), vehicle treated cells; (b), cells incubated with Alexa Fluor 488-conjugated TFPI-2 for 5 min; (c), cells incubated with Alexa Fluor 488-conjugated TFPI-2 for 10 min and (d), cells incubated with Alexa Fluor 488-conjugated TFPI-2 for 2 h. The nucleus is counterstained using DAPI in mounting media. (C), Another set of cells treated with 1 μM TFPI-2 were incubated at 4°C for the indicated times and processed for immunoblotting. For immunoblot analysis, the intensity of blot bands were assessed by densitometric semi-quantitation and depicted by a bar diagram (lower panel). (D), Cells were treated with 50 μg/ml of Alexa Fluor 488-conjugated TFPI-2, incubated at 4°C for the indicated times, and processed for confocal microscopy. (a), vehicle treated cells; (b), cells incubated with Alexa Fluor 488-conjugated TFPI-2 for 5 min; (c), cells incubated with Alexa Fluor 488-conjugated TFPI-2 for 10 min and (d), cells incubated with Alexa Fluor 488-conjugated TFPI-2 for 2 h. At 4°C, Alexa Fluor 488-conjugated TFPI-2 treated cells exhibited a punctate pattern of intracellular TFPI-2 distribution. [Arrows in panel (d) indicates the presence of the protein in the nucleus].
Intracellular distribution of TFPI-2 in endothelial cells and/or cells stably-transfected to express TFPI-2
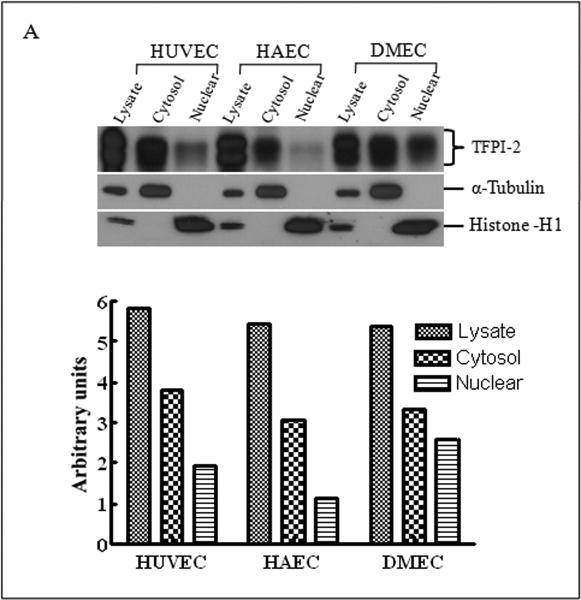
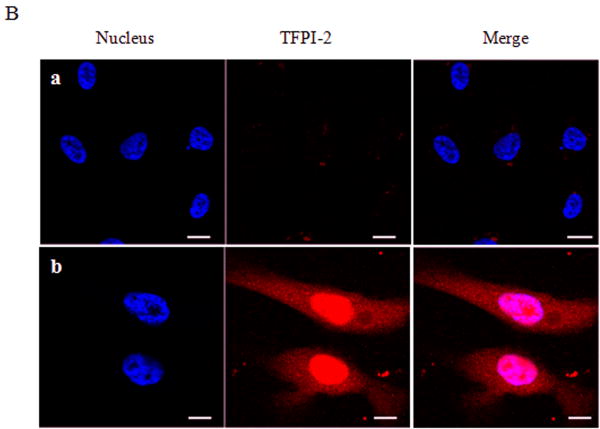
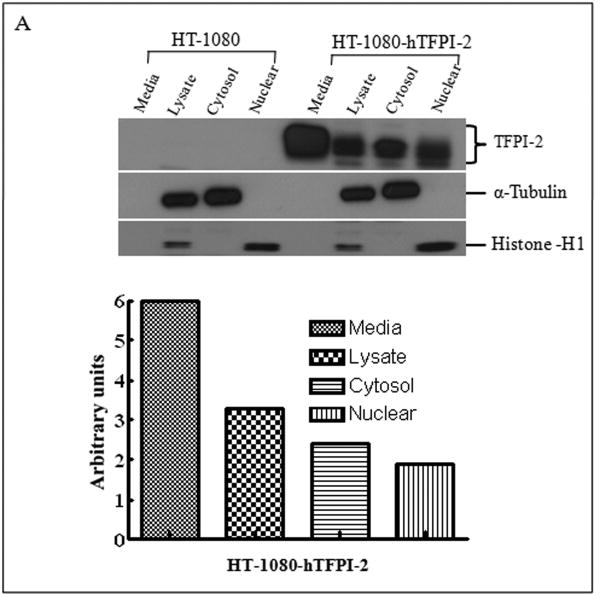
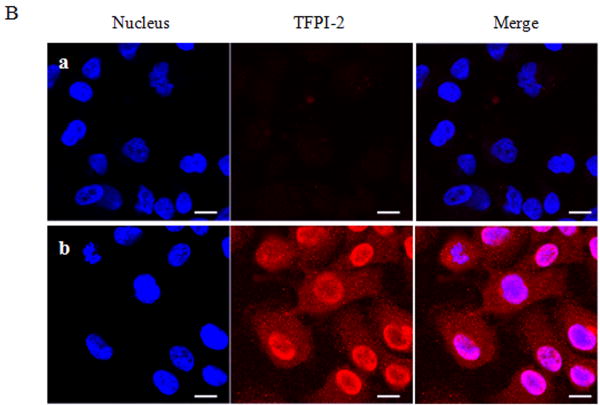
Having demonstrated that exogenous TFPI-2 was internalized by HT-1080 cells that do not synthesize TFPI-2, we next investigated whether TFPI-2 was nuclear-associated in cells that either constitutively synthesize this protein or cells stably-transfected with TFPI-2 cDNA to express this protein. Our initial studies focused on three human endothelial cells lines derived from umbilical vein (HUVEC), aorta (HAEC) and dermal microvascular (DMEC), all known to express varying amounts of TFPI-2 [2]. The results of immunoblotting experiments of endothelial cell fractions clearly demonstrate cytosolic and nuclear localization of TFPI-2 by these cells (Fig. 2A). Immunocytochemistry studies, using a TFPI-2-specific murine monoclonal antibody (SK-9) and Alexa Fluor 555-conjugated goat anti-mouse IgG secondary antibody confirmed the cytosolic and nuclear localization of TFPI-2 in DMECs (Fig. 2B), whereas cells treated exclusively with secondary antibody exhibited no fluorescence, attesting to the specificity of SK-9. Immunoblotting and immunocytochemistry of HT-1080 cells overexpressing TFPI-2 also demonstrated cytosolic and nuclear localization of TFPI-2 in comparison to untransfected cells (Fig. 3). In addition, essentially identical results were obtained with HEK 293 cells overexpressing TFPI-2 (data not shown).
Figure 2. Intracellular distribution of TFPI-2 in endothelial cells assessed by immunoblotting and immunocytochemistry.
To investigate the intracellular distribution of endogenously expressed TFPI-2 protein in endothelial cells, HUVEC, HAEC and DMEC cells were cultured under standard growth conditions. (A), For immunoblotting, lysate and cell fractions from all cells were prepared and probed with anti-TFPI-2 IgG. The purity of cytosolic and nuclear fractions was verified using anti-alpha-tubulin and anti-histone-H1 antibodies, respectively. For immunoblot analysis, the intensity of blot bands were assessed by densitometric semi-quantitation and depicted by a bar diagram (lower panel). (B), DMEC cells were grown on two-chamber culture slides and immunostained with murine monoclonal antibody SK-9 and Alexa Fluor-555-conjugated goat anti-mouse IgG as the secondary antibody. The nucleus is counterstained using DAPI in mounting media. (a), vehicle plus Alexa Fluor-555-conjugated goat anti-mouse IgG. (b), SK-9 antibody and Alexa Fluor-555-conjugate goat anti-mouse IgG.
Figure 3. Analyses of TFPI-2 localization in untransfected and stably-transfected HT-1080 cells by immunoblotting and immunocytochemistry.
Wild-type HT-1080 and HT-1080 cells stably-transfected with hTFPI-2 cDNA were cultured under standard growth conditions. (A), For immunoblotting, total lysate and cell fractions were prepared and probed with anti-TFPI-2 antibody as described earlier. The purity of cytosolic and nuclear fractions was verified using anti-alpha-tubulin and anti-histone-H1 antibodies, respectively. For immunoblot analysis, the intensity of blot bands were assessed by densitometric semi-quantitation and depicted by a bar diagram (lower panel). (B), For immunocytochemistry, cells were grown on two-chamber culture slides and incubated with murine monoclonal antibody SK-9 followed by Alexa Fluor-555-conjugated goat anti-mouse IgG as the secondary antibody. The nucleus is counterstained using DAPI in mounting media. (a), HT-1080 cells (b), HT-1080 cells overexpressing TFPI-2.
Identification of a nuclear localization signal sequence in TFPI-2
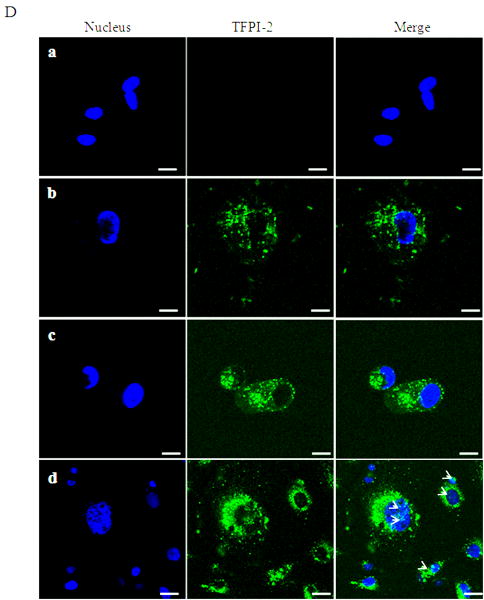
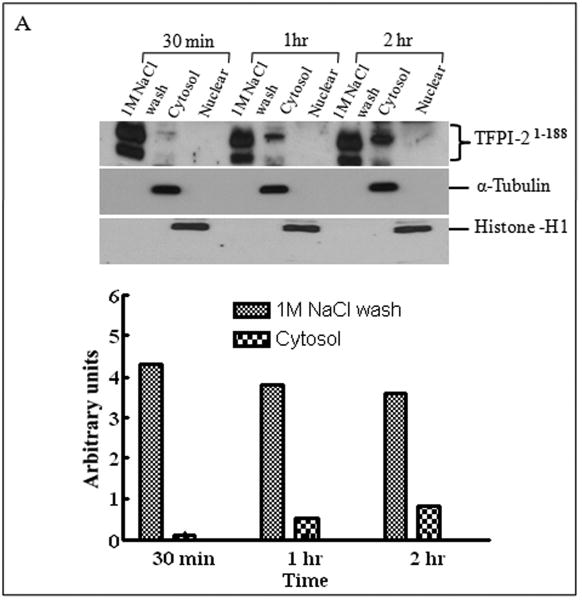
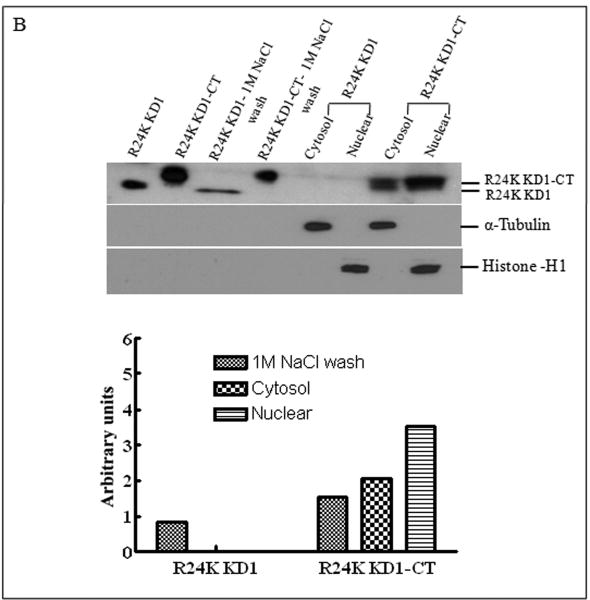
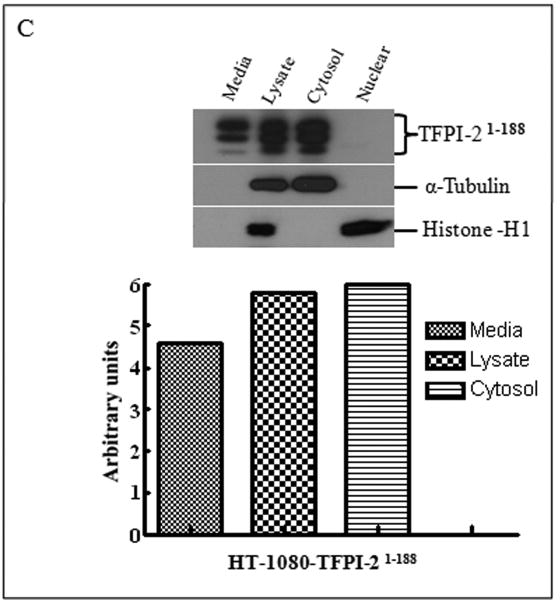
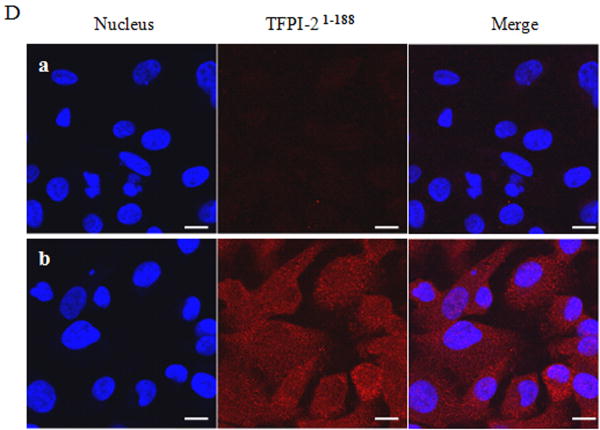
Inasmuch as several internalized or cytosolic proteins are transported to the nucleus by importins that recognize and bind to nuclear localization signal (NLS) sequences in proteins [20], we subjected the TFPI-2 sequence to an in-silico analysis using PredictNLS Imput program (https://cubic.bioc.columbia.edu/predictNLS) to determine if it contained a consensus NLS site. The results of this analysis clearly demonstrated that the carboxy-terminal tail of TFPI-2 contains a putative bipartite NLS sequence (KKKKKMPKLRFASRIRKIRKK) between residues 212-233, which has been experimentally demonstrated in 31 other nuclear-associated proteins. In order to demonstrate the importance of the NLS sequence in the translocation of TFPI-2 to the nucleus, we expressed and purified a TFPI-2 construct (TFPI-21-188) devoid of the C-terminal tail, as well as a chimera of the first Kunitz-type domain and the C-terminal tail (R24K KD1-CT). We then offered these purified proteins to HT-1080 cells and monitored their intracellular trafficking by immunoblot analyses. As shown in Figure 4, TFPI-21-188 was slowly internalized, but failed to be transported to the nucleus (Fig. 4A). While the R24K KD1 failed to enter the cells, the R24K KD1-CT chimera was rapidly internalized and translocated to the nucleus by HT-1080 cells within 5 min (Fig. 4B). In addition, HT-1080 cells stably-transfected with an expression vector bearing the TFPI-21-188 cDNA also failed to demonstrate nuclear localization (Fig. 4C). Immunocytochemical analyses confirmed that TFPI-21-188 was confined to the cytoplasm in the HT-1080 cells overexpressing this protein (Fig. 4D).
Figure 4. Internalization of recombinant R24K KD1-CT and TFPI-21-188 and localization of overexpressing TFPI-21-188 in HT-1080 cells by immunoblotting and immunocytochemistry.
(A), Cells grown in 6 well plates were treated with 1 μM TFPI-21-188 and incubated at 37°C for the indicated times. (B), Cells were treated with either 1 μM R24K KD1 or 1 μM R24K KD1-CT and incubated at 37°C for 5 min. Cells from both (A) and (B) experiments were harvested for fractionation to detect the offered protein. The fractions were probed with anti-TFPI-2 antibody as described in Methods. The purity of cytosolic and nuclear fractions was verified using anti-alpha-tubulin and anti-histone-H1 antibodies, respectively. For immunoblot analysis, the intensity of blot bands were assessed by densitometric semi-quantitation and depicted by a bar diagram (lower panels). (C), Total cell lysate and cell fractions from stably-transfected cells were prepared and probed with anti-TFPI-2 antibody. For immunoblot analysis, the intensity of blot bands were assessed by densitometric semi-quantitation and depicted by a bar diagram (lower panel). (D), The control and TFPI-2 1-188 overexpressing cells were grown on two-chamber culture slides and immunostained with murine monoclonal antibody SK-9 followed by Alexa Fluor 555-conjugated goat anti-mouse IgG as the secondary antibody. The nucleus is counterstained using DAPI in mounting media. (a), HT-1080 cells (b), HT-1080 cells overexpressing TFPI-21-188.
Co-immunoprecipitation of TFPI-2-importin-α complexes from cell lysates
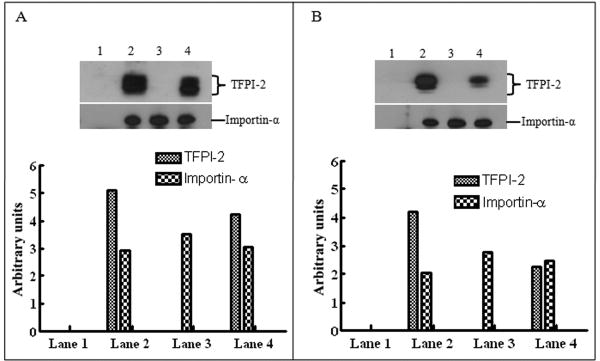
Having demonstrated that TFPI-2 contains a putative bipartite NLS sequence essential for translocation to the nucleus, we next performed co-immunoprecipitation studies to determine whether TFPI-2 was forming complexes with importin-α, a cytosolic protein that binds to NLS-bearing proteins. Using either an anti-TFPI-2 monoclonal antibody (SK-9) or an anti-importin- α monoclonal antibody, each coupled to an insoluble resin, complexes of TFPI-2 and importin-α were immunoprecipitated by each antibody from the cell lysates of either TFPI-2-treated HT-1080 cells or HT-1080 cells overexpressing TFPI-2 (Fig. 5). In contrast, wild-type, untransfected HT-1080 cell lysates immunoprecipitated with SK-9 failed to yield TFPI-2-importin-α complexes, while importin-α was readily immunoprecipitated from all cell lysates by the anti-importin-α antibody (Fig. 5). These results provide definitive evidence that TFPI-2 forms cytosolic complexes with importin-α, which ultimately interacts with importin-β, the karyopherin that chaperones this ternary complex through the nuclear pore complex [21].
Figure 5. TFPI-2 interacts with importin-α.
(A), HT-1080 cells overexpressing TFPI-2 and untransfected cells were subjected to co-immunoprecipitation using SK-9 and importin-α antibodies as described in Methods. The complexes were immunoblotted separately using anti-TFPI-2 and anti-importin-α antibodies. Lane 1, HT-1080 untransfected cell lysate co-immunoprecipitated with SK-9; lane 2, HT-1080 cells overexpressing TFPI-2 co-immunoprecipitated with SK-9; lane 3, HT-1080 untransfected cell lysate co-immunoprecipitated with anti-importin-α; lane 4, HT-1080 cells overexpressing TFPI-2 co-immunoprecipitated with anti-importin-α. For immunoblot analysis, the intensity of blot bands were assessed by densitometric semi-quantitation and depicted by a bar diagram (lower panel). (B), HT-1080 cells were treated with 1 μM TFPI-2 and processed for co-immunoprecipitation. The complexes were similarly immunoblotted using anti-TFPI-2 and anti-importin-α antibodies. Lane 1, lysate from vehicle treated HT-1080 cells co-immunoprecipitated with SK-9; lane 2, lysate from TFPI-2 treated HT-1080 cells co-immunoprecipitated with SK-9; lane 3, lysate from vehicle treated HT-1080 cells co-immunoprecipitated with anti-importin-α; lane 4, lysate from TFPI-2 treated HT-1080 cells co-immunoprecipitated with anti-importin-α. For immunoblot analysis, the intensity of blot bands were assessed by densitometric semi-quantitation and depicted by a bar diagram (lower panel).
Discussion
We previously reported that human TFPI-2, an ECM-associated Kunitz-type serine proteinase inhibitor, induces caspase-mediated apoptosis in several human tumor cell lines when exogenously offered to these cells [8]. In the course of those studies, we observed that TFPI-2 was internalized by the human fibrosarcoma cell line HT-1080, which do not constitutively synthesize this protein [8]. Using immunoblotting and immunocytochemistry approaches, we now report that TFPI-2 is rapidly internalized by HT-1080 cells and efficiently translocated to the nucleus. Translocation of TFPI-2 to the nucleus required a putative bipartite NLS sequence in its carboxy-terminal tail, as a recombinant TFPI-2 construct lacking this domain was slowly internalized by HT-1080 cells but failed to translocate to the nucleus. Moreover, co-immunoprecipitation experiments revealed that TFPI-2 formed complexes with importin-α, a cytosolic protein that recognizes NLS-bearing proteins and, together with importin-β, shuttles these cargo proteins into the nucleus [21]. Interestingly, in addition to TFPI-2-negative cells offered TFPI-2 protein, cells that either constitutively synthesize TFPI-2 or cells that overexpress this protein also contained TFPI-2 in their nuclei, suggesting that a portion of TFPI-2 is internalized by these cells post-secretion.
As TFPI-2 induces apoptosis in several tumor and endothelial cells, the question naturally arises as to whether internalization and nuclear translocation of TFPI-2 are associated with apoptosis induction in these cells. Inasmuch as R24K KD1, a more potent inducer of apoptosis than TFPI-2, failed to be internalized by HT-1080 cells following a 48 h incubation, it would appear that internalization and apoptosis induction may be distinctly different and independent processes. Based on the available data, TFPI-2 localization into the nucleus appears to be a natural phenomenon seen in all cell types that synthesize and express this molecule.
In addition to containing the NLS, the carboxy-terminal tail of TFPI-2 appears to play a significant role in the rapid internalization of TFPI-2 by these cells. In this regard, we observed that a mutant of the first Kunitz-type domain of TFPI-2 (R24K KD1) was not internalized by HT-1080 cells. However, a chimera of this domain and the C-terminal tail, R24K KD1-CT, was rapidly internalized by these cells, providing evidence for the importance of the C-terminal tail in this process. As offered TFPI-2 and R24KD1-CT were each internalized and translocated to the nucleus within minutes, the likelihood that these are internalized through some receptor-mediated endocytic process seems unlikely. Accordingly, the carboxy-terminal tail of TFPI-2, which contains a large number of arginine and lysine residues, appears to be responsible for efficient protein transduction through the plasma membrane, similar to that observed for the Drosophila Antennapedia homeoprotein [22, 23], the HIV-1 TAT protein transduction domain [24], and 12-mers of polylysine and polyarginine [25]. Thus, the C-terminal tail of TFPI-2 may be bifunctional, containing the NLS sequence for nuclear translocation, as well as serving as a protein transduction domain that allows efficient and rapid penetration of the protein through the plasma membrane by a macropinocytotic mechanism. This possible mode of cellular entry does not rule out additional, perhaps receptor-mediated, mechanisms for TFPI-2 internalization since the TFPI-2 construct lacking the C-terminal tail was also internalized by these cells, albeit at a very slow rate in relation to the intact TFPI-2 molecule and the R24K KD1-CT chimera.
TFPI-2 joins a long list of serine proteinase inhibitors previously shown to be localized to the nucleus, which include B-43/PI-6 [9], PI-10 [10], PI-9 [11], PI-8 [12] and MASPIN [13]. These nuclear-associated serpins are intracellular in origin, and all belong to the ovalbumin branch of the serpin superfamily. To our knowledge, TFPI-2 represents the first Kunitz-type serine proteinase inhibitor localized to the nucleus. However, the role of TFPI-2 and the other serpins in the nucleus is unknown. Presumably, these nuclear-associated serine proteinase inhibitors regulate one or more serine proteinases in the nucleus, such as urokinase-type plasminogen activator [26], Omi/HtrA2 [27], granzyme B [28], and other, less characterized nuclear serine proteinases [29,30]. In addition to serine proteinases, two cysteinyl proteinases have been reported in the nucleus that either degrade the small ubiquitin-like modifier [31], or several transcription factors in embryonal carcinoma cells undergoing differentiation [32]. The mechanism whereby these cysteinyl proteinases are regulated in the nucleus is unclear, but one of the nuclear-associated ovalbumin-type serpins, PI-9, has been shown to exhibit cross-class specificity and inhibit a number of cysteinyl proteinases [33]. Unlike TFPI-2 that interacts with serine proteinases non-covalently, serpins form SDS stable covalent complexes with their target proteinase. In this connection, Bird et al. [11] have demonstrated SDS-stable complex formation between PI9 and granzyme B in the nuclear fraction of a human NK leukemia cell line (YT). Whether TFPI-2 can inhibit isolated serine proteinases from HT-1080 nuclear fractions is currently under investigation in our laboratory. In addition, the effect of wild-type and R24Q TFPI-2 on gene expression in HT-1080 cells is also under investigation in our laboratory.
In summary, the studies presented here reveal that human TFPI-2 is rapidly internalized by several different cell lines, and is translocated to the nuclei of these cells. Nuclear translocation of TFPI-2 required a putative NLS sequence in its carboxy-terminal tail, as well as complex formation with importin-α. The functional role of TFPI-2 in the nucleus is unknown, but presumably involves regulation of one or more nuclear-associated serine proteinases.
Acknowledgments
Microscopic images were generated in the University of New Mexico Cancer Center Fluorescence Microscopy Facility, http://hsc.unm.edu/crtc/microscopy/Facility.html.
This work was supported by a National Institutes of Health Research Grant HL64119 (to W.K.).
The abbreviations used are
- TFPI-2
tissue factor pathway inhibitor-2
- KD1
kunitz domain 1
- ECM
extracellular matrix
- DMEM
Dulbecco's minimal essential medium
- PBS
phosphate-buffered saline
- TBS
Tris-buffered saline
- NLS
nuclear localization signal
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Authors' Contributions: P.K. and W.K. designed the experiments; H.S.C made the TFPI-2 constructs; P.K. and W.K. performed the experiments; P.K. and W.K. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rao CN, Peavey CL, Liu YY, Lapiere JC, Woodley DT. Dermatol. 1995;104:379–383. doi: 10.1111/1523-1747.ep12665851. [DOI] [PubMed] [Google Scholar]
- 2.Iino M, Foster DC, Kisiel W. Arterioscler Thromb Vasc Biol. 1998;18:40–46. doi: 10.1161/01.atv.18.1.40. [DOI] [PubMed] [Google Scholar]
- 3.Herman MP, Sukhova GK, Kisiel W, Foster D, Kehry MR, Libby P, Schoenbeck UJ. Clin Invest. 2001;107:1117–1126. doi: 10.1172/JCI10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugiyama T, Ishii S, Yamamoto J, Irie R, Saito K, Otuki T, Wakamatsu A, Suzuki Y, Hio Y, Ota T, Nishikawa T, Sugano S, Masuho Y, Isogai T. FEBS Lett. 2002;517:121–128. doi: 10.1016/s0014-5793(02)02588-7. [DOI] [PubMed] [Google Scholar]
- 5.Chand HS, Foster DC, Kisiel W. Thromb Haemost. 2005;94:1122–1130. doi: 10.1160/TH05-07-0509. [DOI] [PubMed] [Google Scholar]
- 6.Chand HS, Du X, Ma D, Inzunza HD, Kamei S, Foster D, Brodie S, Kisiel W. Blood. 2004;103:1069–1077. doi: 10.1182/blood-2003-06-1930. [DOI] [PubMed] [Google Scholar]
- 7.Rao CN, Mohanam S, Puppala A, Rao JS. Biochem Biophys Res Commun. 1999;255:94–98. doi: 10.1006/bbrc.1999.0153. [DOI] [PubMed] [Google Scholar]
- 8.Kempaiah P, Kisiel W. Apoptosis. 2008;13:702–15. doi: 10.1007/s10495-008-0207-8. [DOI] [PubMed] [Google Scholar]
- 9.Nishibori M, Nakaya N, Ohtsuka A, Murakami T, Saeki K. Histochem Cell Biol. 1998;110:51–56. doi: 10.1007/s004180050264. [DOI] [PubMed] [Google Scholar]
- 10.Chuang TL, Schleef RR. J Biol Chem. 1999;274:11194–11198. doi: 10.1074/jbc.274.16.11194. [DOI] [PubMed] [Google Scholar]
- 11.Bird CH, Blink EJ, Hirst CE, Buzza MS, Steele PM, Sun J, Jans DA, Bird PI. Mol Cell Biol. 2001;21:5396–5407. doi: 10.1128/MCB.21.16.5396-5407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strik MC, Bladergroen BA, Wouters D, Kisiel W, Hooijberg JH, Verlaan AR, Hordijk PL, Schneider P, Hack CE, Kummer JA. J Histochem Cytochem. 2002;50:1443–1453. doi: 10.1177/002215540205001103. [DOI] [PubMed] [Google Scholar]
- 13.Marioni G, Giacomelli L, D'Alessandro E, Marchese-Ragona R, Staffieri C, Ferraro SM, Staffieri A, Blandamura S. Am J Otolaryngol. 2008;29:156–162. doi: 10.1016/j.amjoto.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt AE, Chand HS, Cascio D, Kisiel W, Bajaj SP. J Biol Chem. 2005;280:27832–27838. doi: 10.1074/jbc.M504105200. [DOI] [PubMed] [Google Scholar]
- 15.Du X, Deng FM, Chand HS, Kisiel W. Arch Biochem Biophys. 2003;417:96–104. doi: 10.1016/s0003-9861(03)00332-1. [DOI] [PubMed] [Google Scholar]
- 16.Arepally GM, Kamei S, Park KS, Kamei K, Li ZQ, Liu W, Siegel DL, Kisiel W, Cines DB, Poncz M. Blood. 2000;95:1533–1540. [PubMed] [Google Scholar]
- 17.Sprecher CA, Kisiel W, Mathewes S, Foster DC. Proc Natl Acad Sci USA. 1994;91:3353–3357. doi: 10.1073/pnas.91.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kempaiah P, Chand HS, Kisiel W. Mol Cancer. 2007;6:20. doi: 10.1186/1476-4598-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Pischon N, Palamakumbura AH, Trackman PC. Am J Physiol Cell Physiol. 2007;292:2095–2102. doi: 10.1152/ajpcell.00613.2006. [DOI] [PubMed] [Google Scholar]
- 20.Planque N. Cell Communication and Signaling. 2006;4:7. doi: 10.1186/1478-811X-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Derossi D, Joliot AH, Chassaings G, Prochiantz A. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 23.Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A. J Biol Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 24.Frankel AD, Pabo CO. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 25.Mi Z, Mai J, Lu X, Robbins PD. Mol Therapy. 2000;2:339–347. doi: 10.1006/mthe.2000.0137. [DOI] [PubMed] [Google Scholar]
- 26.Stepanova V, Lebedeva T, Kuo A, Yarovoi S, Tkachuk S, Zaitsev S, Bdeir K, Dumler I, Marks MS, Parfyonova Y, Tkachuk VA, Higazi AA, Cines DB. Blood. 2008;112:100–110. doi: 10.1182/blood-2007-07-104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuninaka S, Iida SI, Hara T, Nomura M, Naoe H, Morisaki T, Nitta M, Arima Y, Mimori T, Yonehara S, Saya H. Oncogene. 2007;26:2395–2406. doi: 10.1038/sj.onc.1210042. [DOI] [PubMed] [Google Scholar]
- 28.Trapani JA, Browne KA, Smyth MJ, Jans DA. J Biol Chem. 1996;271:4127–4133. doi: 10.1074/jbc.271.8.4127. [DOI] [PubMed] [Google Scholar]
- 29.Chen HJ, Hwong CL, Wang CH, Hwang J. J Biol Chem. 2000;275:13109–13117. doi: 10.1074/jbc.275.17.13109. [DOI] [PubMed] [Google Scholar]
- 30.Meyer J, Jucker M, Ostertag W, Stocking C. Blood. 1998;91:1901–1908. [PubMed] [Google Scholar]
- 31.Bailey D, O'Hare P. J Biol Chem. 2004;279:692–703. doi: 10.1074/jbc.M306195200. [DOI] [PubMed] [Google Scholar]
- 32.Scholtz B, Lamb K, Rosfjord E, Kingsley M, Rizzino A. Dev Biol. 1996;173:420–42. doi: 10.1006/dbio.1996.0037. [DOI] [PubMed] [Google Scholar]
- 33.Annand RR, Dahlen JR, Sprecher CA, Foster DC, Mankovich J, Talanian R, Kisiel W, Giegel DA. Biochem J. 1999;342:655–665. [PMC free article] [PubMed] [Google Scholar]