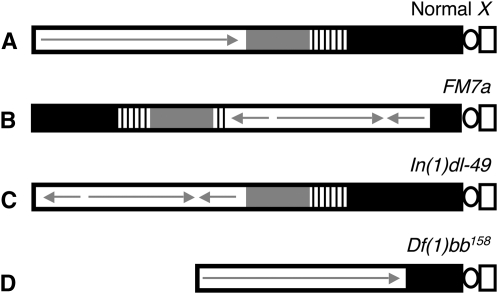
Figure 3.—
Diagram illustrating the different X chromosomes used in NDJ tests. (A) A normal X chromosome is shown with the centromeric constriction on the right. Euchromatin is depicted as a white rectangle, with a continuous arrow indicating a lack of rearrangements. The shaded and striped regions represent heterochromatin with the 11-Mb satellite DNA (359-bp repeat) (solid) and rDNA (stripes). (B) The FM7a balancer chromosome contains multiple inversions within the euchromatin (represented with arrows) as well as a rearrangement that places a large region of centromere-proximal heterochromatin at the distal end of the chromosome. (C) The In(1)dl-49 chromosome contains an inversion in the euchromatin that reduces recombination to ∼20% of wild type. However, centromere-proximal heterochromatin is unaffected. (D) The Df(1)bb158 contains a deletion that removes ∼80% of the centromere-proximal heterochromatin but the euchromatic region of the chromosome is normal.