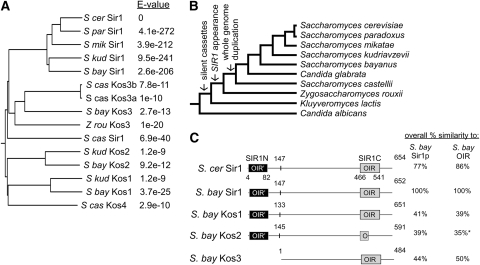
Figure 1.—
SIR1 family represented a rapidly evolving family of paralogs that diverged after the whole-genomewide duplication. (A) N-J bootstrap tree of SIR1 paralogs with E-value of alignment with S. cerevisiae Sir1. (B) Evolutionary tree of several Saccharomyces species and other yeast (Wolfe 2006). (C) Representation of S. cerevisiae Sir1 protein and paralogs from S. bayanus. Sir1 protein from S. bayanus was aligned with Sir1 from S. cerevisiae and with paralogous sequences from S. bayanus, called Kos1–3 (Kin of Sir1). The OIR is boxed. The amino-terminal duplication of the OIR is represented by a solid box labeled OIR′. Amino acid similarity to the full-length S. bayanus Sir1 protein and to its OIR was determined by BLAST. *Kos2 contained significant gaps in the OIR alignment compared to S. cerevisiae OIR. The similarity to S. bayanus OIR was based only on the partial alignment.