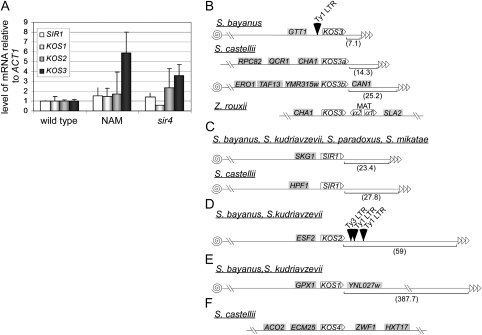
Figure 7.—
Telomere-position-effect regulation of, and genomic organization of, SIR1 and KOS paralogs in Saccharomyces and Zygosaccharomyces. (A) Transcriptional regulation of SIR1 and KOS genes in S. bayanus. Levels of mRNA of SIR1 paralogs were measured by QRT–PCR from cells treated with nicotinamide (NAM) or from cells containing a sir4 mutation relative to ACT1 mRNA and then normalized to wild-type cells. (B–F) Chromosomes are depicted as a line, with telomeres as triangles and centromeres as spirals. On the basis of genes encoded on the same contig and using sequence from S. cerevisiae genome, each paralog's distance in kilobases from the telomeres was predicted as indicated in parentheses below the chromosome. The positions of S. cerevisiae Ty elements are labeled above the chromosome as triangles. The names of each flanking gene is in a shaded box. (B) Predicted locations of KOS3 paralogs in S. bayanus, S. castellii, and Z. rouxii. (C) Predicted locations of SIR1 in S. bayanus, S. kudriavzevii, S. paradoxus, S. mikatae, and S. castellii. (D) Predicted locations of KOS2 paralogs in S. bayanus and S. kudriavzevii. (E) Predicted locations of KOS1 paralogs in S. bayanus and S. kudriavzevii. (F) Predicted locations of KOS4 in S. castellii.