Abstract
In generating a conditional transgenic murine model based on a tetracycline-regulated system, we obtained unexpected patterns of expression due to the transcriptional inactivity of the tet-responder promoter. Here we show strong cell-type-restricted expression that was variegated to an extent determined by the number of responder transgene copies integrated into the host genome.
THE tetracycline-based tet-off system (Gossen and Bujard 1992) has emerged as a powerful tool for consistent and conditional induction of transcription of transgenic sequences (Bockamp et al. 2002). This has been exploited to generate transgenic organisms, including models for the study of pathological mechanisms involved in human diseases, such as nervous system targets (Mayford et al. 1996; Tremblay et al. 1998; Yamamoto et al. 2000; Lucas et al. 2001; SantaCruz et al. 2005). In this context, we planned a tet-conditional murine model (Figure 1A) to dissect out the chain of events related to the G93A amino-acid substitution in the human superoxide dismutase type 1 (SOD1) protein sequence (Gurney et al. 1994), which leads to disease in a subset of familial amyotrophic lateral sclerosis.
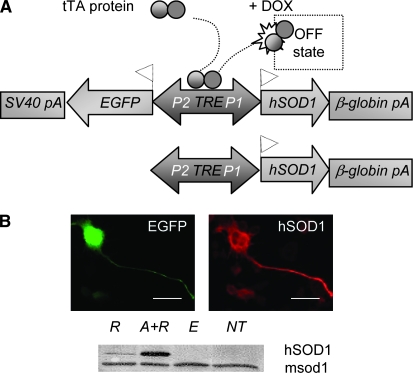
Figure 1.—
Experimental settings. (A) The tTA binds the core promoter (TRE; tetracycline-responder element) and activates bidirectional expression (P1, P2; minimal promoters of cytomegalovirus). Antibiotic administration (DOX; the tetracycline derivative doxycycline) abolishes transcriptional activation (OFF state). The cDNA coding for a mutated version of human SOD1 (hSOD1) was cloned into the pBI-EGFP vector (Clontech, Palo Alto, CA) and two linear constructs—4.4 kb (top: EGFP-TRE-hG93ASOD1cDNA; 997, 995, 497 transgenic lines) and 2.4 kb (bottom: TRE-hG93ASOD1cDNA; 450, 446, 428, 419, 413 transgenic lines)—were generated and micro-injected into BDF1 (C57BL/6xDBA/2) fertilized oocytes. The activator mouse line PrP-tTA F959 in the FVB/N strain (Tremblay et al. 1998) was crossed to each responder transgenic line to obtain double-transgenic mice. (B) Transiently transfected NS34 cells (green, EGFP; red, monoclonal anti-human SOD1 antibody; MBL International, Woburn, MA). A representative immunoblot is shown. Bar, 50 μm. R, responder plasmid alone; A+R, activator and responder plasmid; E, empty responder vector; NT, nontransfected cells; msod1, murine superoxide dismutase type I.
Restricted cell-type expression in vivo for both enhanced green fluorescent protein and human SOD1 products:
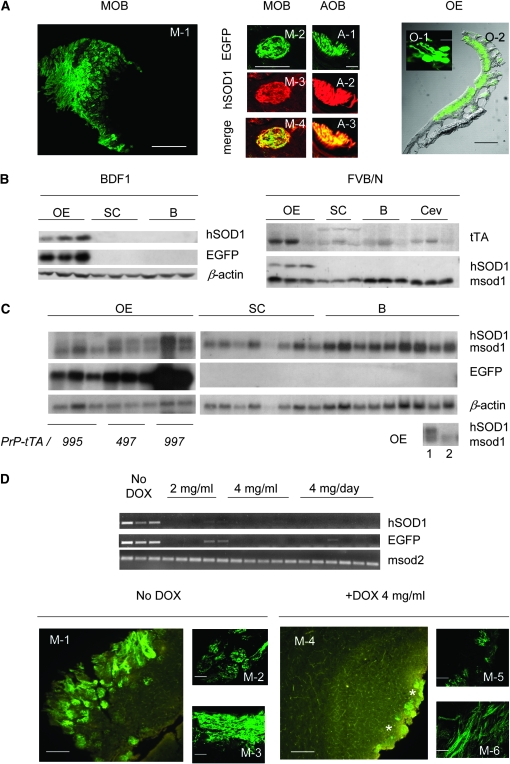
We observed a powerful induction of human SOD1 and enhanced green fluorescent protein (EGFP) expression in the transiently transfected NSC-34 motor-neuron-like cell line (Figure 1B). This confirmed that our tet-off-based system was working in a cell-culture context (Babetto et al. 2005). However, when eight independent responder transgenic mouse lines were generated, both EGFP and human SOD1 products were restricted to some incoming projections toward the main and accessory olfactory bulbs in the double-transgenic mice (MOB and AOB in Figure 2A). An extended analysis of the olfactory mucosa revealed that the reporter EGFP signal could be ascribed exclusively to the mature olfactory receptor neurons (ORNs) (Figure 2A). In contrast, there was no evidence of exogenous protein expression in the remaining cells of the olfactory epithelium or in the central nervous system (data not shown). This restricted pattern was observed in all of the scored animals (38 double-transgenic mice) along all of the transgenic lines, except on a single occasion (data not shown), and it was confirmed by Western blotting (Figure 2B). Analysis of total RNA samples revealed that the human SOD1 and EGFP mRNAs were present exclusively in extracts from the olfactory epithelium (OE; Figure 2C). This observation strongly indicated that the responder promoter could be inactive at the transcriptional level, therefore leading to a lack of protein synthesis. However, the system showed fully functional behavior in terms of doxycycline response in the expressing cells (Figure 2D). Therefore, the ability to turn off expression of both EGFP and human SOD1 in the ORNs after administration of doxycycline demonstrated that in vivo the expressed tetracycline-controlled transactivator (tTA) protein retained all of its functional features.
Figure 2.—
Restricted patterns of expression and doxycycline dependence. (A) Representative pattern of expression in tissue sections obtained by transcardial perfusion of double-transgenic mice (two to five animals/line). Main olfactory bulbs (MOB, M-1: bar, 800 μm; M-2–M-4: bar, 100 μm) and accessory olfactory bulbs (AOB, A-1–A-3: bar, 25 μm). Olfactory receptor neurons (O-1: bar, 10 μm) and olfactory epithelium (OE, O-2: bar, 500 μm). (B) Representative immunoblots [anti-EGFP monoclonal (Roche, Manheim, Germany); anti-SOD1 (Upstate, Lake Placid, NY); and monoclonal anti-β-actin (Immunological Sciences, Rome)]. (Left) Homogenates from double-transgenic mice from a representative EGFP-TRE-hG93ASOD1cDNA responder line in the BDF1 mouse strain background (tTA immunoblot in supplemental information). (Right) The same situation after changing the background of the same line to FVB/N (up to the eighth generation). (C) Northern blots (two to three double-transgenic mice from the three EGFP-TRE-hG93ASOD1cDNA lines). (Inset bottom right) Controls: 1, double-transgenic mouse; 2, nontransgenic mouse. (D) RT–PCR and representative tissue sections of doxycycline-treated mice dosed as indicated in drinking water with 5% sucrose over 1 week, compared to nontreated (No DOX) counterparts (three to six mice/group). The glomerular layer in the main olfactory bulb (M-1 and M-4: bar, 200 μm; M-2 and M-5: bar, 100 μm) and axonal projections (M-3 and M-6: bar, 50 μm) are shown. Stars indicate weak and low numbers of autofluorescent glomeruli. OE, olfactory epithelium; SC, spinal cord; B, brain; Cev, cerebellum; msod1 and msod2, murine superoxide dismutase types I and II.
The efficiency of transgene expression in olfactory receptor neurons is inversely correlated with the number of responder transgene copies inserted:
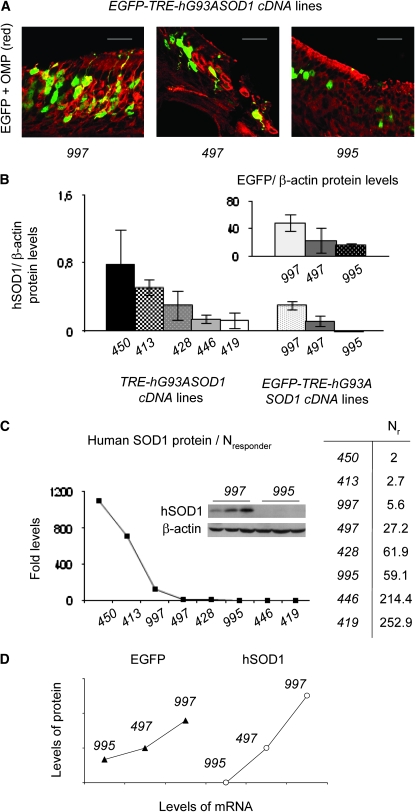
Immunohistochemical analysis of the OE showed that expression of EGFP in the ORNs was variegated (Figure 3A), which could be related to the levels of the transgenic proteins in OE homogenates across the transgenic lines (Figure 3B). The levels of production of both the human SOD1 and EGFP were inversely proportional to the number of transgene responder copies (Figure 3C), irrespective of the linear construct tested (Figure 3B). This trend is consistent with the repeat-induced transgene-silencing theory (Garrick et al. 1998) observed in constitutively expressing transgenes. The presence of a major number of transgene copies has been associated with a major chance of epigenetic repression (Manuelidis 1991; Jones and Takai 2001). Thus, in our mice, the responder sequences might favor the attraction of repressing states in concomitance with an increase in copy numbers, leading to a minor number of expressing ORNs.
Figure 3.—
Degree of copy-number-dependent expression variegation in the olfactory epithelium. (A) EGFP (green) signal in OMP-stained (red) sections of olfactory epithelium of EGFP-TRE-hG93ASOD1cDNA double-transgenic mice. Bars: 20 μm (997 and 497) and 30 μm (995). (B) Quantification of protein immunoblots from olfactory epithelium extracts (three to five mice/transgenic line), normalized to β-actin. (C) Efficiency of human SOD1 expression (fold levels). Protein levels normalized to β-actin and the number of responder transgene copies [Nresponder (Nr), inset right] estimated by real-time PCR and Southern blotting (data not shown). Inset center: immunoblot showing decrease of hSOD1 production when responder copies increase only 10-fold. (D) Correlation between mean mRNA and protein levels based on Western and Northern blots (Figure 1C and data not shown).
The responder transgene sequences as the cause of the unexpected expression pattern:
There is documented evidence that random integration of transgenes can fail to drive consistent exogenous expression due to several factors, such as genome position influences (Sabl and Henikoff 1996), mouse genetic background (Opsahl et al. 2002; Padjen et al. 2005) and number (Saveliev et al. 2003) and orientation (Stam et al. 1998) or sequence composition (Ramírez et al. 2001) of transgene-item repeats, especially if viral sequences are included (Schumacher et al. 2000). Failure of tet-based strategies in vivo has also been reported. This has been related either to defective tTA expression (Böger and Gruss 1999; Fedorov et al. 2001; Lee et al. 2006) or to epigenetic repression on the tet-promoter activity (Janicki et al. 2004; Pankiewicz et al. 2005; Kues et al. 2006), in addition to undesired interactions of eukaryotic cell factors with the tet-promoter sequences (Rang and Will 2000; Gould and Chernajovsky 2004).
In our mice, the genomic insertion context had little influence on the pattern of distribution observed for EGFP/human SOD1 expression, since the same distribution patterns occurred in eight different responder transgenic lines (data not shown). Neither qualitative nor quantitative tTA defects can explain the expression patterns displayed by our transgenic lines (supplemental information and data not shown). Therefore, consistent data support the ubiquitous production of tTA and the effective transcriptional activation of tet promoters (Boy et al. 2006) when other responder mouse lines are crossed to the same PrP-tTA activator line used in this study. Moreover, the unexpected pattern observed in our mice persisted after changing some responder lines to the FVB/N strain background (Figure 2B and data not shown).
The lack of EGFP and/or human SOD1 mRNAs can be interpreted as a consequence of the absence of transcriptional activation of the responder promoter in almost all cell types. The proportional trend between human SOD1 and EGFP production (Figure 3, B and D) suggests that the system works bidirectionally (Krestel et al. 2001) and reinforces the hypothesis of transcriptional inactivity since the EGFP and human SOD1 coding sequences share the same transcriptional link. In addition, the striking correspondence between mRNA and protein levels supports this impression (Figure 3D). Otherwise, it must be assumed that post-transcriptional regulation affected each independent element separately and along all of the transgenic lines. The expression variegation observed at the ORNs, and especially the inverse dependence of expression levels on responder transgene copy numbers, could be indirectly indicative of repressing states affecting the responder sequences and might also be influenced by the composition of our transgene sequences. This is dramatically underlined by having obtained the same outcome in all of the responder transgenic lines that were generated. In conclusion, we raise concerns about the composition of the responder transgene sequences because the aberrant patterns of transgene expression can be dramatic when these murine models are used to study human diseases, a concern that can be extended to in vivo conditional RNA-interference-related technologies.
Acknowledgments
The authors thank C. P. Berrie for editorial assistance; B. A. Whitelaw and R. Giovannoni for critical reading; S. B. Prusiner and F. L. Margolis for the gift of the PrP-tTA mice and the anti-OMP serum; and C. Bendotti, L. Conforti, M. M. Barzago, and M. Rizzi. Procedures regarding the mice were in compliance with international laws and policies. This work was supported by the Italian Telethon Foundation.
References
- Babetto, E., A. Mangolini, M. Rizzardini, M. Lupi, L. Conforti et al., 2005. Tetracycline-regulated gene expression in the NSC-34-tTA cell line for investigation of motor neuron diseases. Brain Res. Mol. Brain Res. 140 63–72. [DOI] [PubMed] [Google Scholar]
- Bockamp, E., M. Maringer, C. Spangenberg, S. Fees, S. Fraser et al., 2002. Of mice and models: improved animal models for biomedical research. Physiol. Genomics 11 115–132. [DOI] [PubMed] [Google Scholar]
- Böger, H., and P. Gruss, 1999. Functional determinants for the tetracycline-dependent transactivator tTA in transgenic mouse embryos. Mech. Dev. 83 141–153. [DOI] [PubMed] [Google Scholar]
- Boy, J., T. V. Leergaard, T. Schmidt, F. Odeh, U. Bichelmeier et al., 2006. Expression mapping of tetracycline-responsive prion protein promoter: digital atlasing for generating cell-specific disease models. Neuroimage 33 449–462. [DOI] [PubMed] [Google Scholar]
- Fedorov, L. M., O. Y. Tyrsin, O. Sakk, A. Ganscher and U. R. Rapp, 2001. Generation dependent reduction of tTA expression in double transgenic NZL-2/tTACMV mice. Genesis 31 78–84. [DOI] [PubMed] [Google Scholar]
- Garrick, D., S. Fiering, D. I. K. Martin and E. Whitelaw, 1998. Repeat-induced gene silencing in mammals. Nat. Genet. 18 56–59. [DOI] [PubMed] [Google Scholar]
- Gossen, M., and H. Bujard, 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, D. J., and Y. Chernajovsky, 2004. Endogenous GATA factors bind the core sequence of the tetO and influence gene regulation with the tetracycline system. Mol. Ther. 10 127–138. [DOI] [PubMed] [Google Scholar]
- Gurney, M. E., H. Pu, A. Y. Chiu, M. C. Dal Canto, C. Y. Polchow et al., 1994. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264 1772–1775. [DOI] [PubMed] [Google Scholar]
- Janicki, S. M., T. Tsukamoto, S. E. Salghetti, W. P. Tansey, R. Sachidanandam et al., 2004. From silencing to gene expression: real-time analysis in single cells. Cell 116 683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, P. A., and D. Takai, 2001. The role of DNA methylation in mammalian epigenetics. Science 293 1068–1070. [DOI] [PubMed] [Google Scholar]
- Krestel, H. E., M. Mayford, P. H. Seeburg and R. Sprengel, 2001. A GFP-equipped bidirectional expression module well suited for monitoring tetracycline regulated gene expression in mouse. Nucleic Acids Res. 29 E39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kues, W. A., R. Schwinzer, D. Wirth, E. Verhoeyen, E. Lemme, et al., 2006. Epigenetic silencing and tissue independent expression of a novel tetracycline inducible system in double-transgenic pigs. FASEB J. 20 1201–1202. [DOI] [PubMed] [Google Scholar]
- Lee, S., R. Agah, M. Xiao, A. D. Frutkin, M. Kremen et al., 2006. In vivo expression of a conditional TGF-beta1 transgene expression in SM22alpha-tTA transgenic mice. J. Mol. Cell. Cardiol. 40 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, J. J., F. Hernández, P. Gómez-Ramos, M. A. Morán, R. Hen et al., 2001. Decreased nuclear beta-catenin tau hyperphosphorylation and neurodegeneration in GSK-3beta conditional transgenic mice. EMBO J. 20 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis, L., 1991. Heterochromatic features of an 11-megabase transgene in brain cells. Proc. Natl. Acad. Sci. USA 88 1049–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford, M., M. E. Bach, Y-Y. Huang, L. Wang, R. D. Hawkins et al., 1996. Control of memory formation through regulated expression of a CaMKII transgene. Science 274 1678–1683. [DOI] [PubMed] [Google Scholar]
- Opsahl, M. L., M. McClenaghan, A. Springbett, S. Reid, R. Lathe et al., 2002. Multiple effects of genetic background on variegated transgene expression in mice. Genetics 160 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padjen, K., S. Ratnam and U. Storb, 2005. DNA methylation precedes chromatin modifications under the influence of the strain-specific modifier Ssm1. Mol. Cell. Biol. 25 4782–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiewicz, R., Y. Karlen, M. O. Imhof and N. Mermod, 2005. Reversal of the silencing of tetracycline-controlled genes requires the coordinate action of distinctly acting transcription factors. J. Gene Med. 7 117–132. [DOI] [PubMed] [Google Scholar]
- Ramírez, A., E. Milot, I. Ponsa, C. Marcos-Gutiérrez, A. Page et al., 2001. Sequence and chromosomal context effects on variegated expression of keratin 5/lacZ constructs in stratified epithelia of transgenic mice. Genetics 158 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rang, A., and H. Will, 2000. The tetracycline-responsive promoter contains functional interferon-inducible response elements. Nucleic Acids Res. 28 1120–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabl, J. F., and S. Henikoff, 1996. Copy number and orientation determine the susceptibility of a gene to silencing by nearby heterochromatin in Drosophila. Genetics 142 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaCruz, K., J. Lewis, T. Spire, J. Paulson, L. Kotilinek et al., 2005. Tau suppression in a neurodegenerative mouse model improves memory function. Science 309 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveliev, A., C. Everett, T. Sharpe, Z. Webster and R. Festenstein, 2003. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature 422 909–913. [DOI] [PubMed] [Google Scholar]
- Schumacher, A., P. A. Koetsier, J. Hertz and W. Doerfler, 2000. Epigenetic and genotype-specific effects on the stability of de novo imposed methylation patterns in transgenic mice. J. Biol. Chem. 275 37915–37921. [DOI] [PubMed] [Google Scholar]
- Stam, M., A. Viterbo, J. N. M. Mol and J. M. Kooter, 1998. Position-dependent methylation and transcriptional silencing of transgenes in inverted T-DNA repeats: implications for posttranscriptional silencing of homologous host genes in plants. Mol. Cell. Biol. 18 6165–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay, P., Z. Meiner, M. Galou, C. Heinrich, C. Petromilli et al., 1998. Doxycycline control of prion protein transgene expression modulates prion disease in mice. Proc. Natl. Acad. Sci. USA 95 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., J. J. Lucas and R. Hen, 2000. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell 101 57–66. [DOI] [PubMed] [Google Scholar]