Abstract
In Arabidopsis, SHOOT MERISTEMLESS (STM) and CLAVATA1 (CLV1) competitively regulate meristem homeostasis. Here, we explore the interaction of their maize homologs knotted1 (kn1) and thick tassel dwarf1 (td1). kn1 mutants form fewer lateral organs and td1 inflorescences are fasciated with additional floral organs. Double mutants show kn1 epistatic to td1 in seedling and ear development but dose-sensitivity exists later to promote leaf initiation. Thus kn1 and td1 function in a pathway to maintain meristem homeostasis but their products may interact with different partners during development.
TO produce organs predictably, meristems tightly control the opposing processes of meristem maintenance and lateral organ initiation. In Arabidopsis, the CLAVATA (CLV) genes control balance between these processes. Loss-of-function/hypomorphic clv mutants have enlarged shoot and floral meristems, producing more floral organs (Leyser and Furner 1992; Clark et al. 1993, 1995; Kayes and Clark 1998). CLV1 and CLV2 encode a leucine-rich repeat (LRR) receptor-like kinase protein and an LRR protein lacking a kinase domain, respectively (Clark et al. 1997; Jeong et al. 1999). CLV3 encodes a small peptide (Fletcher et al. 1999) that physically interacts with CLV1 (Ogawa et al. 2008). BAM1, BAM2, and BAM3 encode CLV1-related receptor kinases. Opposite of clv mutants, the double or triple bam mutants have smaller meristems (DeYoung et al. 2006). Genetic evidence, however, points to a significant role for the BAM loci in the CLV pathway, as the bam mutants ameliorate the clv3 phenotype but enhance null alleles of clv1 (DeYoung and Clark 2008).
The homeobox gene SHOOT MERISTEMLESS (STM), central to meristem maintenance and determinacy (Long et al. 1996), functions in a separate pathway (Brand et al. 2002; Lenhard et al. 2002). Strong stm mutants lack vegetative development and produce only cotyledons (Barton and Poethig 1993). Weak stm mutants may progress to flowering and demonstrate a function for STM in inflorescences (Endrizzi et al. 1996; Brand et al. 2002; Bhatt et al. 2004; Kanrar et al. 2006). clv1 alleles suppress the stm phenotype and stm alleles suppress the clv1 phenotype, indicating that these genes play antagonistic roles. This genetic interaction is dose dependent: CLV1 and STM proteins are sensitive to each other, suggesting that their balance is necessary to maintain proper meristem size and shape (Clark et al. 1996).
Knotted1 (kn1) is a maize homolog of STM with similar expression pattern and function (Smith et al. 1992; Vollbrecht et al. 2000). Expressivity of recessive kn1 mutations depends upon genetic background and meristem size. In inbred lines with larger meristems, defects are most pronounced during the adult phase: tassels are less branched, ears often absent or with reduced seed set. Ectopic leaves may form above the ear node (Kerstetter et al. 1997). In restrictive backgrounds with smaller meristems, a limited shoot phenotype is seen in which zero to two leaves form (Vollbrecht et al. 2000).
clv-like mutations affect distinct meristems in the grasses, unlike Arabidopsis, in which all shoot meristems are larger. Mutations in the CLV3 rice homolog, FON2, affect only floral meristems (Suzaki et al. 2006), while mutations in another CLV3 homolog, FCP1, affect only vegetative meristems (Suzaki et al. 2008). Mutations in the rice CLV1 homolog, FON1, affect only floral meristems (Suzaki et al. 2004), suggesting another CLV1 homolog functions in inflorescence and vegetative meristems. thick tassel dwarf (td1) encodes the most similar maize CLV1 homolog with 58% amino acid identity to CLV1 and 51% identity to BAM1 and BAM2. td1 mutants have enlarged inflorescence meristems, increased spikelet density and supernumerary floral organs but, in contrast, reduced vegetative growth. Plants are shorter with fewer leaves (Bommert et al. 2005). Thus TD1 may promote vegetative meristem growth and restrict inflorescence and floral meristem growth.
To investigate the role of STM- and CLV1-homologous pathways in the grasses, we have utilized mutations in kn1 and td1 to observe double-mutant phenotypes.
td1-glf fails to suppress the limited shoot phenotype of kn1-E1:
We combined the null kn1-E1 allele (Vollbrecht et al. 2000) with the null td1-glf allele (Bommert et al. 2005) to determine if td1-glf can suppress the limited shoot phenotype caused by kn1-E1. The limited shoot phenotype was more penetrant in a mixed B73:Mo17 background than in B73 alone, allowing us to study more individuals. Of 39 kn1-E1 homozygous plants, 8 (20.5%) exhibited limited shoots (Table 1; Figure 1A). The genotype at the td1 locus did not affect this phenotype, indicating that td1-glf cannot suppress the effects of kn1-E1 during early vegetative growth and that kn1-E1 is epistatic to td1-glf.
TABLE 1.
Penetrance of limited shoot phenotype in the B73:Mo17 genetic background
| Genotypea | N | LSb (n) |
|---|---|---|
| kn1/kn1; td1/td1 | 18 | 3c |
| kn1/kn1; td1/+ | 14 | 4 |
| kn1/kn1; +/+ | 7 | 1c |
| kn1/+; td1/td1 | 11 | 0 |
| kn1/+; td1/+ | 13 | 0 |
| kn1/+; +/+ | 5 | 0 |
| Total | 68 | 8 |
To create a family segregating double mutants in the B73:Mo17 background, kn1-E1 was backcrossed to Mo17 four times and td1-glf was backcrossed to B73 nine times. An F1 plant with these two parents was self-pollinated to make an F2 family and assayed by PCR for all possible allelic combinations. Only kn1-E1 plants showed the limited shoot phenotype. An F2 individual with the genotype kn1-E1/+ td1-glf/+ was crossed to a sibling with the genotype kn1-E1/kn1-E1 td1-glf/+ to enrich for double mutants. Plants were grown under greenhouse conditions.
Not significantly different from expected ratio (X2, P > 0.01).
Limited shoot phenotype.
One plant in each of these classes recovered and grew normally.
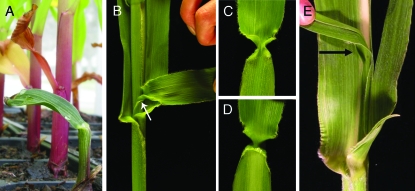
Figure 1.—
Vegetative phenotypes of double mutants. (A) A kn1-E1; td1-glf seedling showing the limited shoot phenotype of a single leaf. Normal siblings are in the background. Their first leaf has senesced. (B) An ectopic leaf in the axil of a normal leaf. The ligule (arrow) is facing the ligule of the normal leaf (not seen). The ectopic leaf is attached at the meristem. (C–D) Each surface of ectopic leaf from B. (E) A fused ectopic leaf from a double mutant, fused proximal to arrow.
td1-glf vegetative meristems are smaller:
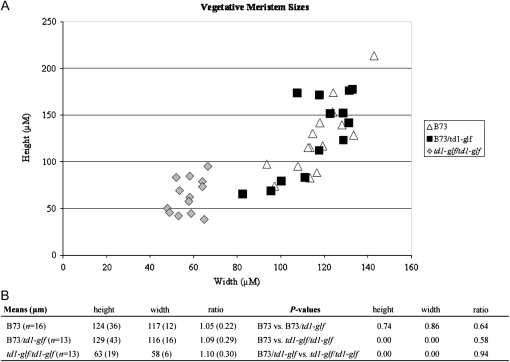
In Arabidopsis, clv1 meristems are larger than wild type during vegetative growth (Clark et al. 1993). In contrast, we found that vegetative meristems of td1-glf mutants are smaller than those of wild-type siblings (Figure 2). This finding may explain why td1-glf does not suppress kn1-E1.
Figure 2.—
td1-glf vegetative meristems are smaller. (A) Height and width (micrometers) of B73 (open triangles), B73/td1-glf (solid squares), and td1-glf/td1-glf (shaded diamonds) were measured and plotted. (B) Mean heights, widths, and ratios for each genetic class. Standard deviations are in parentheses. P-values for t-tests comparing the classes are also shown. The lack of statistical difference in ratios for the three classes indicates they are proportional. B73 is statistically similar to B73/td1-glf heterozygotes, indicating that a single td1-glf allele has no affect on vegetative meristem size. Plants were grown in a growth chamber with the following conditions: 16 hr of light at 26°; 8 hr of darkness at 22°. Vegetative meristems were measured 14 days after sowing (DAS) after dissection and viewing under a Nikon SMZ800 microscope.
td1-glf increases the penetrance of the kn1-E1 ectopic vegetative leaf phenotype:
In permissive backgrounds, plants homozygous for kn1 loss-of-function alleles develop ectopic leaves in the axils of leaves at a low penetrance (12%) (Kerstetter et al. 1997). td1-glf enhanced this phenotype dose dependently (Table 2) (Figure 1, B–E). The ectopic leaves are reversed in polarity with their adaxial surface (Figure 1D) facing the adaxial surface of the true leaf. In an F2 family segregating td1-glf and kn1-E1, all double mutants had at least one ectopic leaf, while kn1-E1 homozygotes with one or no copies of td1-glf had 40% and 20%, respectively. In a family segregating 50% kn1-E1 and 50% kn1-E1/+ in the td1-glf mutant background, 90% of double mutants and 40% of kn1-E1 heterozygotes had ectopic leaves. No kn1-E1/+ td1-glf/td1-glf plants had ectopic leaves in the F2 family, indicating possible epigenetic and/or environmental influence on the phenotype. We also observed fusion of most ectopic leaves of double mutants to the adjacent leaf (Figure 1E) whereas fusion only occurred in a quarter of ectopic leaves of kn1-E1/+ td1-glf/td1-glf plants. Thus, we detected a synergistic increase in penetrance of ectopic leaves and an unexpected involvement of td1 in the regulation of leaf initiation.
TABLE 2.
td1-glf increases the penetrance of the kn1-E1 ectopic leaf phenotype and promotes fusion of ectopic leaves in the B73 genetic background
| Genotypea | Ectopic leavesb | Penetrance (%) |
|---|---|---|
| F2 family | ||
| kn1/kn1; td1/td1 | 4 (2.5; 4) | 100 |
| kn1/kn1; td1/+ | 4 (0.7; 10) | 40 |
| kn1/kn1; +/+ | 1 (0.2; 5) | 20 |
| kn1/+; td1/td1 | 0 (0; 13) | 0 |
| kn1/+; td1/+ | 0 (0; 19) | 0 |
| kn1/+; +/+ | 0 (0; 19) | 0 |
| +/+; td1/td1 | 0 (0; 11) | 0 |
| +/+; td1/+ | 0 (0; 5) | 0 |
| +/+; +/+ | 0 (0; 10) | 0 |
| 1:1 family | ||
| kn1/kn1; td1/td1 | 9 (1.90; 10) | 90 (12/19)c |
| kn1/+; td1/td1 | 8 (0.45; 20) | 40 (2/9) |
td1-glf was backcrossed to B73 nine times and the kn1-E1 allele was backcrossed to B73 six times. An F1 plant with these two parents was self-pollinated. F2 plants were assayed by PCR for all possible allelic combinations and a double mutant was crossed with a kn1-E1/+ td1-glf/ td1-glf plant to generate a family segregating 1:1 for kn1-E1 homozygotes in the td1-glf mutant background. Plants were grown under greenhouse conditions.
Not significantly different from expected F2 ratio (X2, P > 0.05).
Number of plants with ectopic leaves followed by mean number of ectopic leaves per plant and sample size in parentheses.
Number of fused ectopic leaves/total number of ectopic leaves.
kn1-E1 is epistatic to td1-glf in ear development:
kn1 and td1 are antagonistic in ear development. In permissive backgrounds, ears of kn1 mutants are small with reduced spikelet density (Kerstetter et al. 1997). Ears of td1 mutants are fasciated with increased spikelet density on a continuous meristematic surface (Figure 3B) (Bommert et al. 2005). Double-mutant ears (Figure 3D) resembled those of kn1 plants (Figure 3C), with patches of rachis lacking spikelets and similar spikelet density (Table 3). However, instead of solid rachis, ectopic spikelets formed inside ears of double mutants (Figure 3E) and the rachis fused distally, a phenotype not observed in kn1 or td1 single-mutant ears (data not shown).
Figure 3.—
Reproductive phenotypes. Unpollinated ears of (A) normal sibling, (B), td1-glf homozygote, (C) kn1-E1 homozygote, and (D) a double mutant. Patches of cob have failed to initiate kernel primordia in the double mutant, as seen in kn1-E1 homozygotes. (E) A longitudinal section of the same ear in D. Arrow in E points to ectopic kernel primordia in the ear core, a phenotype not seen in either single mutant.
TABLE 3.
kn1-E1 is epistatic to td1-glf in ear spikelet formation
| Genotype | Ear spikeletsa (n) |
|---|---|
| A. Ear spikelet counts | |
| +/−; +/− | 25.39 (12.91; 31) |
| td1/td1; +/− | 30.90 (16.14; 23) |
| +/−; kn1/kn1 | 10.89 (5.82; 11) |
| td1/td1; kn1/kn1 | 11.67 (5.01; 6) |
| Genotype | P-values |
| B. Comparisons of ear spikelet counts of the td1-glf; kn1-E1 F2 family | |
| td1/td1; +/− vs. +/−; +/− | 0.002** |
| +/−; kn1/kn1 vs. +/−; +/− | 0.000*** |
| td1/td1; +/− vs. +/−; kn1/kn1 | 0.000*** |
| td1/td1; kn1/kn1 vs. +/−; +/− | 0.001** |
| td1/td1; kn1/kn1 vs. td1/td1; +/− | 0.000*** |
| td1/td1; kn1/kn1 vs. +/−; kn1/kn1 | 0.975 |
Plants from the td1-glf; kn1-E1 F2 family were grown in the greenhouse and observed. (A) Unpollinated ear spikelet counts. Numbers are mean values with standard deviations in parentheses followed by the number of plants measured. (B) P-values of a two-tailed Student's t-test of ear spikelet counts comparing the genetic classes. **Values are significant at the 1% level; ***values are significant at the 0.1% level.
Numbers of spikelets were counted from a cross section of the ear at its widest point, ∼1 cm from the ear tip.
Summary:
STM and CLV loci competitively regulate the balance of central and peripheral zones of the shoot apical meristem (Clark et al. 1996). To assess whether this relationship exists in maize, we analyzed the interaction of td1 and kn1. td1-glf does not suppress the limited shoot phenotype of kn1-E1, possibly because td1 vegetative meristems are smaller. kn1-E1 suppresses the fasciation of td1-glf inflorescences, indicating that the abnormal growth of fasciated meristems requires normal kn1 function. td1-glf fails to suppress the ectopic leaves of kn1 mutants; rather, it enhances the phenotype. td1-glf increases fusion of these ectopic leaves to their subtending leaves, a defect ameliorated by normal kn1 alleles. In addition, a new phenotype was seen in double mutants: ectopic spikelets inside ears. Thus, KN1 and TD1 function in a linear pathway maintaining homeostasis in the vegetative meristem and ear, during spikelet initiation. Because td1 mutant ears are fasciated yet their vegetative meristems are smaller, the function of TD1 is similar to CLV1 in the inflorescence and floral meristems, but more similar to BAM (DeYoung et al. 2006) in vegetative meristems. The synergism displayed by ectopic leaves and ectopic spikelets suggests that TD1 and KN1 also converge to maintain cob identity and lateral organ initiation. Like the BAM genes, td1 is widely expressed (Bommert et al. 2005) and these multiple roles may be dependent upon tissue specificity of other pathway members. We conclude that the relationship between td1 and kn1 is not directly comparable to that of CLV1 and STM, as td1, like the BAM genes, has multiple functions throughout development.
Acknowledgments
We thank Rebecca Bart, Rachel Bond, Future Zhou, Laticia Holley, and Lan Ma for their assistance in fieldwork, data collection, and analysis. Members of the Hake lab provided helpful critique of the manuscript. David Hantz and Julie Calfas provided excellent plant care. Funding for this project was from National Science Foundation DBI 0604923 grant (to S.H.).
References
- Barton, M. K., and R. S. Poethig, 1993. Formation of the shoot apical meristem In Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119 823–831. [Google Scholar]
- Bhatt, A. M., J. P. Etchells, C. Canales, A. Lagodienko and H. D Ickinson, 2004. VAAMANA: a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Genes Dev. 328 103–111. [DOI] [PubMed] [Google Scholar]
- Bommert, P. B., C. Lunde, J. Nardmann, E. Vollbrecht, M. P. Running et al., 2005. thick tassel dwarf1 encodes a putative maize orthologue of the Arabidopsis CLAVATA1 leucine-rich receptor-like kinase. Development 132 1235–1245. [DOI] [PubMed] [Google Scholar]
- Brand, U., M. Grunewald, M. Hobe and R. Simon, 2002. Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S. E., M. P. Running and E. M. Meyerowitz, 1993. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S. E., M. P. Running and E. M. Meyerowitz, 1995. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121 2057–2067. [Google Scholar]
- Clark, S. E., S. E. Jacobsen, J. Z. Levin and E. M. Meyerowitz, 1996. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122 1567–1575. [DOI] [PubMed] [Google Scholar]
- Clark, S. E., R. W. Williams and E. M. Meyerowitz, 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 575–585. [DOI] [PubMed] [Google Scholar]
- DeYoung, B. J., K. L. Bickle, K. J. Schrage, P. Muskett, K. Patel et al., 2006. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45 1–16. [DOI] [PubMed] [Google Scholar]
- DeYoung, B. J., and S. E. Clark, 2008. BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi, K., B. Moussian, A. Haecker, J. Z. Levin and T. Laux, 1996. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10 967–979. [DOI] [PubMed] [Google Scholar]
- Fletcher, J. C., U. Brand, M. P. Running, R. Simon and E. M. Meyerowitz, 1999. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis meristems. Science 183 1911–1914. [DOI] [PubMed] [Google Scholar]
- Jeong, S., A. E. Trotochaud and S. E. Clark, 1999. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11 1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanrar, S., O. Onguka and H. M. Smith, 2006. Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 224 2263–2273. [DOI] [PubMed] [Google Scholar]
- Kayes, J. M., and S. E. Clark, 1998. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125 3843–3851. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R. A., D. Laudencia-Chingcuanco, L. G. Smith and S. Hake, 1997. Loss of function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124 3045–3054. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., G. Jurgens and T. Laux, 2002. The WUSHEL and SHOOT MERISTEMLESS genes fulfill complementary roles in Arabidopsis shoot meristem regulation. Development 129 3195–3206. [DOI] [PubMed] [Google Scholar]
- Leyser, H. M. O., and I. J. Furner, 1992. Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116 397–403. [Google Scholar]
- Long, J. A., E. I. Moan, J. I. Medford and M. K. Barton, 1996. A member of the KNOTTED class of homeodomain proteins encoded by the SHOOTMERISTEMLESS gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., H. Shinohara, Y. Sakagami and Y. Matsubayashi, 2008. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319 294. [DOI] [PubMed] [Google Scholar]
- Smith, L. G., B. Greene, B. Veit and S. Hake, 1992. A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116 21–30. [DOI] [PubMed] [Google Scholar]
- Suzaki, T., M. Sato, M. Ashikari, M. Miyoshi, Y. Nagato et al., 2004. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131 5649–5657. [DOI] [PubMed] [Google Scholar]
- Suzaki, T., T. Toriba, M. Fujimoto, N. Tsutsumi, H. Kitano et al., 2006. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 47 1591–1602. [DOI] [PubMed] [Google Scholar]
- Suzaki, T., A. Yoshida and H. Hirano, 2008. Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice. Plant Cell 20 2049–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht, E., L. Reiser and S. Hake, 2000. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127 3161–3172. [DOI] [PubMed] [Google Scholar]