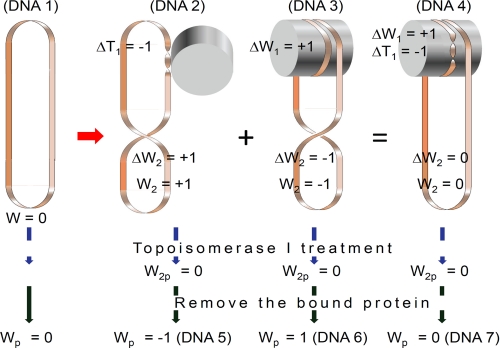
FIGURE 1.
Topological consequences of cc-dsDNA binding to a protein and a model for the binding of cc-dsDNA to Mhr1. The extension and associated untwisting of cc-dsDNA that are required for homologous pairing are induced by RecA/Rad51 binding (29) and generate positive (or left-handed) supercoils in the remaining part of the cc-dsDNA (DNA 2). This type of untwisting is observed as the formation of negatively supercoiled cc-dsDNA by the topoisomerase I treatment of cc-dsDNA bound to RecA/Rad51, followed by the removal of proteins (DNA 5). When the substrate cc-dsDNA for homologous pairing is negatively supercoiled, the positive supercoils that are generated during the reaction are neutralized, and thus, the negative supercoils stimulate homologous pairing by RecA (11). On the other hand, the right-handed wrapping around a protein generates negative (or right-handed) supercoils (DNA 3). This is observed as the generation of positively supercoiled cc-dsDNA, when the cc-dsDNA bound to human Rad52 is treated with topoisomerase I and the proteins are removed (DNA 6) (41). The positive supercoils generated by untwisting can be canceled by the negative supercoils generated by the right-handed wrapping (DNA 4). If the substrate cc-dsDNA is negatively supercoiled, then the negative supercoils prevent right-handed wrapping. These phenomena explain our finding that the negative supercoils of cc-dsDNA prevent homologous pairing by Mhr1 (Figs. 3, 4, 6, and 7) and that the treatment of the cc-dsDNA-Mhr1 complex with topoisomerase generates neither positive nor negative supercoils, as shown in Fig. 8B. The cancellation of the positive supercoils and the negative supercoils is not necessarily complete. In the case of the Rad52-cc-dsDNA complex, the negative supercoils generated by the wrapping are larger than the positive supercoils generated by the untwisting. This figure represents the change by a single unit of supercoil or twist, caused by the binding of originally relaxed cc-dsDNA (DNA 1) to a protein for simplicity. W, writhing number of the entire cc-dsDNA; Wn, local writhing number; ΔWn, local changes in writhe (positive values, left-handed supercoiling or right-handed wrapping); ΔTn, local change in twist (negative values, untwisting) in region n. Subscripts 1 and 2 indicate the protein-bound region of the DNA molecule and the remaining region of the DNA (where ΔT2 = 0), respectively. The subscript p indicates the values after topoisomerase treatment. Before topoisomerase treatment, the sum of the local changes in writhe and twist is zero, because there is no change in the linking number. If the substrate dsDNA is relaxed (W = 0), then W2 is equal to ΔW2. The treatment of the protein-DNA complex by topoisomerase I results in the relaxing of W2 (to become 0). After the removal of proteins, Wp, which is the sum of W1 and W2 (equal to 0), is equal to the sum of ΔT1 and ΔW1.