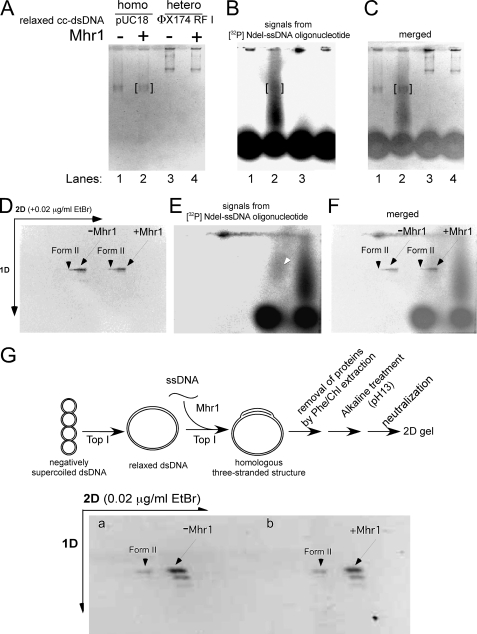
FIGURE 5.
Gel analysis of the protein-free products of homologous three-stranded structure formation by Mhr1 in the presence of calf thymus topoisomerase I. Negatively supercoiled cc-dsDNA (pUC18 and φX174 RF I) was treated with calf thymus topoisomerase I at 37 °C for 30 min. To initiate Mhr1-catalyzed homologous pairing, the relaxed cc-dsDNA and active calf thymus topoisomerase I were added to the reaction mixture containing Mhr1-[32P]NdeI ssDNA oligonucleotide complexes. The solution of homologous (pUC18; 49.0 μm) or heterologous (φX174 RF I; 34.5 μm) relaxed cc-dsDNA was incubated with the reaction mixture of [32P]NdeI ssDNA oligonucleotide (1.3 μm, calculated by assuming that the yield of each 32P labeling step was 100%), Mhr1 (0 and 3.9 μm), and calf thymus topoisomerase I at 37 °C for 30 min. After the proteins were removed by treatments with SDS and proteinase K, the DNA products were analyzed by electrophoresis on 1% agarose gels (2 V/cm) for 11 h in a cold room (4 °C). The second dimension of gel electrophoresis was performed in the presence of 0.02 μg/ml ethidium bromide. After electrophoresis, the gel was dried and exposed to an imaging plate, which was analyzed with a Fuji BAS2000 image analyzer. Due to the limitations of our Radio Isotope facility, it was not possible to obtain images in A and D with the same quality as those in G and Fig. 6. A, the negative print of the first dimensional gel image after ethidium bromide staining. Lanes 1 and 2, homologous combinations in the absence and presence of Mhr1, respectively. Lanes 3 and 4, heterologous combinations in the absence and presence of Mhr1, respectively. B, homologous pairing products represented by the 32P-labeled NdeI-ssDNA oligonucleotide in the same gel shown in A, as detected by exposure to an imaging plate. The positions of pUC18 cc-dsDNA are framed in red. The [32P]NdeI-ssDNA oligonucleotide gradually and continuously dissociated from the homologous pairing products during electrophoresis but formed a distinct signal from the unreacted or free 32P-labeled NdeI-ssDNA oligonucleotide. C, a merged image of A and B. D, the negative print of the second dimensional gel image after ethidium bromide staining. Nicked circular dsDNA (Form II) and cc-dsDNA are indicated by arrowheads and arrows, respectively. -Mhr1, control without Mhr1; +Mhr1, the products from the complete system for the formation of a homologous three-stranded structure by Mhr1 in the presence of the topoisomerase. E, homologous pairing products represented by the 32P-labeled NdeI-ssDNA oligonucleotide in the same gel shown in D, as detected by exposure to an imaging plate. Most of the 32P-labeled NdeI-ssDNA oligonucleotide dissociated from the homologous pairing products but formed a distinct set of signals from a dotlike signal of unreacted 32P-labeled NdeI-ssDNA oligonucleotide. Only a small fraction of the 32P-labeled NdeI-ssDNA oligonucleotide formed a signal at the position of cc-dsDNA, indicated by an arrow. F, a merged image of D and E. G, the negative print showing the topological state of the products of homologous pairing by Mhr1. Homologous (pUC18) relaxed cc-dsDNA (41.8 μm) was incubated with 1.3 μm unlabeled NdeI ssDNA oligonucleotide, Mhr1 (0 and 3.9 μm), and calf thymus topoisomerase I at 37 °C for 30 min. After a phenol/chloroform extraction, the DNA products were subjected to alkaline treatment (pH, 13.0) to dissociate all of the ssDNA oligonucleotide from the cc-dsDNA. After the DNA solution was neutralized to recover the double helix, the cc-dsDNA molecules were analyzed by two-dimensional electrophoresis. The second dimension of gel electrophoresis was performed in the presence of 0.02 μg/ml ethidium bromide. The outline of the experiment is illustrated above the gel image. Nicked circular dsDNA and cc-dsDNA are indicated with arrowheads and arrows, respectively. 1D, first dimension; 2D, second dimension.