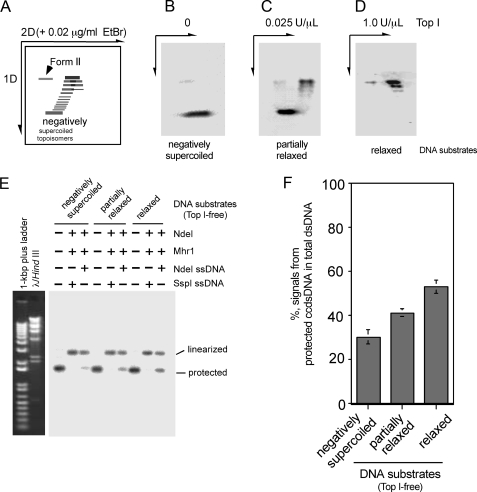
FIGURE 6.
Comparison of negatively supercoiled cc-dsDNA and relaxed cc-dsDNA as the dsDNA substrate for Mhr1-catalyzed homologous three-stranded structure formation in the absence of topoisomerase. A–D, two-dimensional (2D) gel electrophoretic profiles of negatively supercoiled and relaxed pUC18 DNA species. Full or partially relaxed pUC18 cc-dsDNA was prepared by treating supercoiled pUC18 cc-dsDNA with calf thymus topoisomerase I (Top 1), and purifying it from the proteins. The DNA products were analyzed by electrophoresis on 1% agarose gels, and then electrophoresis was performed in the second dimension in the presence of 0.02 μg/ml ethidium bromide. A, schematic diagram. Relaxed cc-dsDNA moves together with nicked circular dsDNA (Form II) in the first dimension (1D) and moves faster than form II DNA in the second dimension. B, the negative print of untreated negatively supercoiled pUC18 after ethidium bromide staining. C, the negative print of partially relaxed pUC18 after ethidium bromide staining. D, the negative print of fully relaxed pUC18 after ethidium bromide staining. E, Mhr1-catalyzed homologous three-stranded structure formation with various dsDNA substrates and ssDNA oligonucleotides. Fully or partially relaxed or negatively supercoiled pUC18 cc-dsDNA (41.8 μm) was incubated with 1.3 μm NdeI ssDNA oligonucleotides or SspI ssDNA oligonucleotides and Mhr1 (0 or 3.9 μm) at 37 °C for 30 min (in the absence of topoisomerase) in the order described in the legend to Fig. 3B. After the incubation, the DNA products were digested with NdeI, and the proteins were removed by SDS and proteinase K treatments. The DNA products were then subjected to electrophoresis on 1% agarose gels containing ethidium bromide. The signals representing the DNA species in the gel were detected by a Southern blot analysis, using 32P-labeled pUC18 as a probe. F, a quantitative representation of the results shown in E. The percentages of the signals representing the DNA species detected by a Southern blot analysis among those from the total dsDNA are plotted, as described in the legend to Fig. 3C.