FIGURE 1.
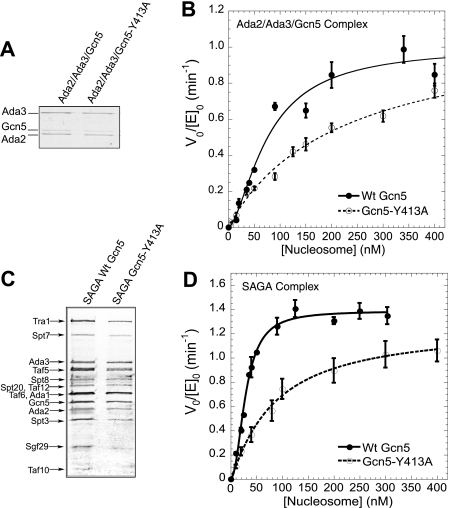
Cooperative nucleosome acetylation by the SAGA complex requires the Gcn5 bromodomain. A, characterization of recombinantly expressed yeast Ada2-Ada3-Gcn5 subcomplex containing either wild-type Gcn5 or Gcn5 with a point mutation in the bromodomain (Gcn5-Y413A). The purified complexes were analyzed by 15% SDS-PAGE and stained with Coomassie Blue. B, the initial velocity of nucleosomal array acetylation per enzyme as a function of nucleosome concentration for Ada2-Ada3-Gcn5 (solid circles) and Ada2-Ada3-Gcn5-Y413A subcomplex (open circles). At least three independent experiments were performed at each nucleosome array concentration. The average initial velocity data were fit to a cooperativity saturation model to give a cooperativity constant of 1.63 ± 0.17 for wild-type Ada2-Ada3-Gcn5 subcomplex (solid line) and 0.91 ± 0.15 for Ada2-Ada3-Gcn5-Y413A subcomplex (dashed line). For direct comparison with data in Fig. 1D, only data for nucleosome concentrations from 0 to 400 nm are shown. The full range of nucleosome concentrations is shown in supplemental Fig. S1. C, characterization of TAP-purified SAGA complex containing either wild-type Gcn5 or Gcn5-Y413A. Subunits were resolved on a gradient gel and visualized by silver staining. Known protein components were labeled according to Winston and co-workers (24). D, the initial velocity of nucleosomal array acetylation per enzyme as a function of nucleosome concentration for wild-type SAGA (solid circles) and SAGA Gcn5-Y413A (open circles). Fitting of the data gives a cooperativity constant of 1.97 ± 0.15 (wild-type SAGA, solid line) and 1.01 ± 0.22 (SAGA Gcn5-Y413A, dashed line). Wild-type data were adapted from Li and Shogren-Knaak (17).