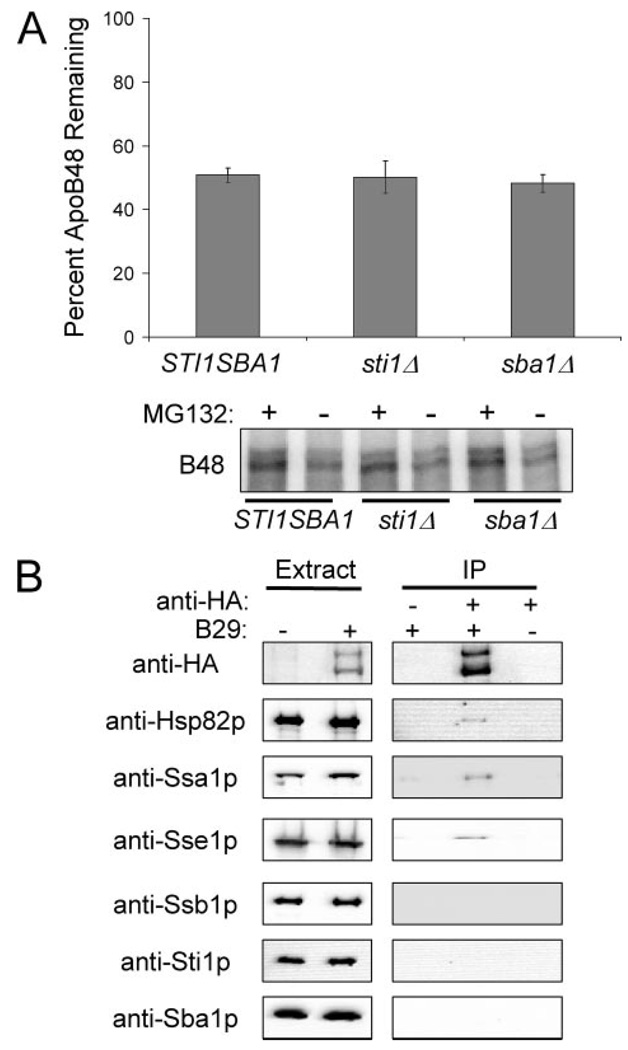
FIGURE 5. The Hsp90 co-chaperones, Sba1p and Sti1p, do not contribute to ApoB ERAD.
A, apoB48 degradation was assessed in vitro at 37 °C for 30 min using cytosol (5 µg/µl) from STI1SBA1, sti1Δ, or sba1Δ yeast. The percentage of apoB remaining was calculated by normalizing the amount of apoB in the Me2SO-treated samples to those samples treated with MG132, thus reflecting only the degree of proteasome-mediated degradation. Data represent the means from six independent experiments ± S.E. of the means. The lower panel contains a representative phosphorimage of 35S-labeled apoB48 during the degradation assay in cytosol from STI1SBA1, sti1Δ, or sba1Δ yeast. Reactions were treated with the proteasome inhibitor MG132 (250 µm) or Me2SO (−), as indicated. B, apoB29 co-precipitates with Hsp82p, Ssa1p, and Sse1p, but not with Ssb1p, Sti1p, or Sba1p. Extracts were prepared from cells transformed with a vector control or with pSLW1-B29 (B29) and were treated with anti-HA resin or an unconjugated Sepharose resin control. The proteins in the precipitates were resolved by SDS-PAGE and were immunoblotted with the indicated antisera.