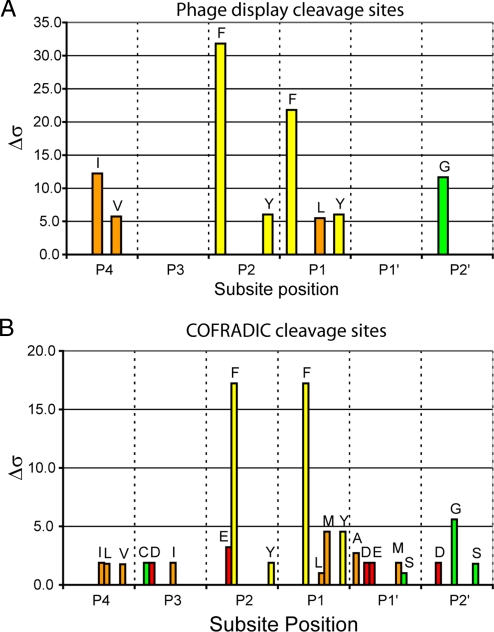
Fig. 5.
Extended substrate specificity of gzmC. (A) A random nonamer phage display library was subject to 4 rounds of selection with 200 nM gzmC in two separate experiments. Sixty-eight phage sequences randomly selected from both experiments were aligned, and amino acid selectivity at each subsite was analyzed with a Δσ cutoff of 5 applied. (B) The P5–P5′ sequence of 19 gzmC cleavage events identified by proteomics was subject to the same analysis as in A except that the Δσ cutoff was reduced to 1 because of the small sample size.