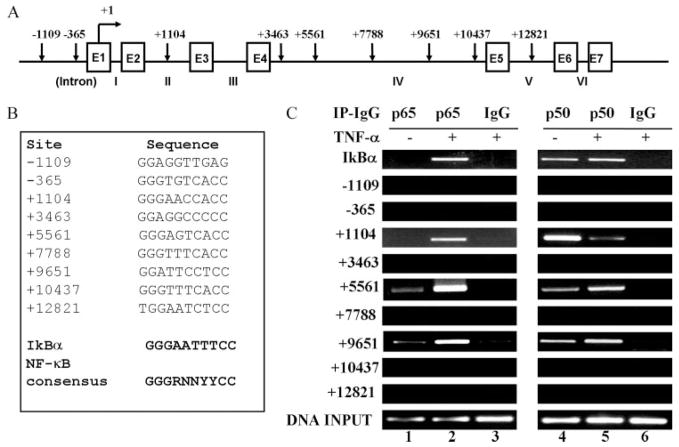
FIGURE 4.
Mapping of NF-κB binding sequence(s) in the human FcRn gene by ChIP. A, Organization of the human FcRn gene. The exon/intron organization of the 15-kb FcRn gene is schematically shown. The positions of the exons/introns and the base counts are shown (GenBank accession no. AC010619). Positions of the putative NF-κB binding sequences relevant to the transcription start site of the FcRn gene are shown at the top. Transcriptional start sites are shown (17). Arrows indicate the position of candidate NF-κB sequences. Exons (E) are drawn as boxes and intron numbers are shown as roman numerals. B, The putative NF-κB binding sequences in the FcRn gene are listed. Numbers represent the putative NF-κB binding sequences relevant to the transcription start site of the FcRn gene. The consensus NF-κB sequence is bolded. R is an A or G purine and Y is a C or T pyrimidine. The NF-κB binding sequence in the promoter of IκB is used as a positive control. C, NF-κB p65 and p50 components are present at FcRn introns in vivo in response to TNF-α. THP-1 cells were treated with TNF-α (50 ng/ml) for 30 min. ChIP assays were performed using p65-specific (lanes 1–3) and p50-specific (lanes 4–6) Abs. IgG was used as a negative control (lanes 3 and 6). Immunoprecipitated chromatin was prepared and subjected to PCR analysis using primer pairs (Table I). ChIP was performed at least three times. IP, Immunoprecipitation.