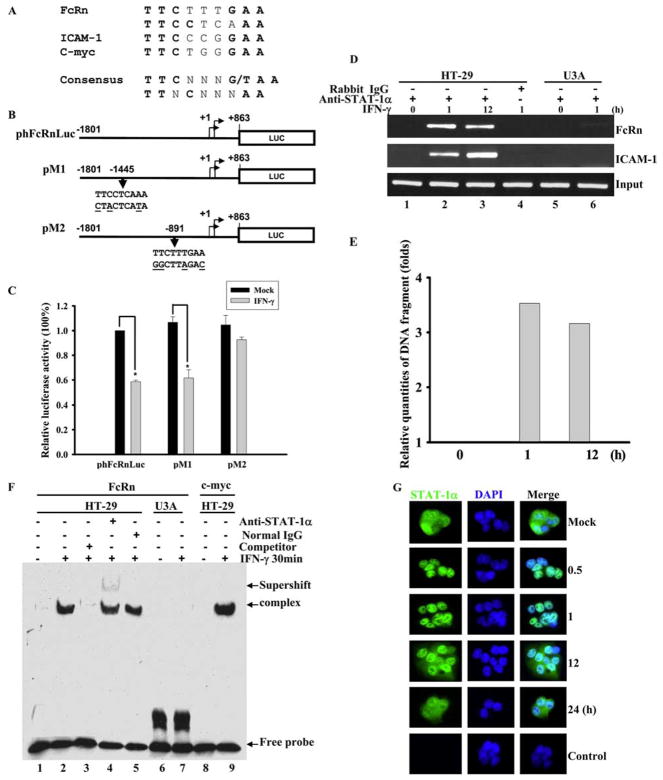
FIGURE 4.
Identification of IFN-γ responsive element in human FcRn promoter. A and B, The putative STAT-1 binding sequences in FcRn gene promoter. A, STAT-1 binding sequences in the promoter of ICAM1 and c-myc were used as a positive control. The consensus STAT-1 sequence is in boldface. N represents any nucleotide. B, A schematic representation of the luciferase reporter constructs. The positions of the base count are shown (GenBank accession no. AC010619). Reporter construct phFcRnLuc contains the FcRn promoter sequence from −1801 to +863 kb. The putative GAS mutations (underlined bases) in constructs pM1 and pM2 are also shown. Arrowheads indicate the position of the STAT-1 binding site in relation to the transcription start site of the FcRn gene. Luc, Luciferase. C, Identification of GAS sequence in response to IFN-γ stimulation. Wild-type 2fTGH cells were transiently transfected with phFcRnLuc, pM1, and pM2 constructs. Twenty-four hours after transfection, cells were either mock-treated (filled bar) or treated with IFN-γ (open bar) for 4 h. Cells were then harvested and protein extracts were prepared for the luciferase assay as described in Materials and Methods. Luciferase activity was measured and normalized to Renilla luciferase content. The results show the mean value from three independent experiments. *, p < 0.05. D and E, Detection of the in vivo binding of STAT-1 protein to the human FcRn promoter in a ChIP assay. D, Formaldehyde-crosslinked chromatin was prepared from both mock-treated and IFN-γ-treated HT-29 (lanes 1– 4) or STAT-1-null U3A (lanes 5 and 6) cells as described in Materials and Methods. ChIP assays were performed using STAT-1-specific Abs (lanes 1–3, 5, and 6) or isotype-matched IgG (lane 4) as a negative control. Immunoprecipitated chromatin was subjected to PCR analysis using FcRn and ICAM-1 specific primers. The equivalent amount of chromatin in the immunoprecipitations was monitored by PCR amplification of input chromatin as an internal control. ChIP assay was performed at least three times. E, Quantitative real-time RT-PCR analysis of chromatin immunoprecipitated PCR products for FcRn at the indicated times. F, EMSA analysis of binding activities of DNA probe with nuclear extracts from HT-29 cells treated with (+) or without (−) IFN-γ. DNA binding was performed using a DNA probe of human FcRn promoter with nuclear extracts from HT-29 or U3A cells treated with or without IFN-γ (25 ng/ml) for 30 min. A 26-bp fragment spanning the putative STAT-1 binding sequence corresponding to the GAS was used as a biotin-labeled probe. Binding specificity of these complexes was examined by competition assays with a 100-fold molar excess of unlabeled STAT-1-specific probe (lane 3). Supershift experiments were performed in the presence of the STAT-1 Ab, resulting in the formation of a slow migrating supershift band (lane 4). Free-labeled probes are also indicated. G, Immunofluorescence images of STAT-1 cellular localization at the indicated times after exposure to IFN-γ (25 ng/ml). HT-29 cells stimulated with IFN-γ were stained with Alexa Fluor 458-labeled-STAT-1-specific Ab, and translocation of STAT-1 into the nucleus was detected by immunofluorescence microscopy as described in Materials and Methods. For correlation of the STAT-1 protein (green) with nuclear morphology, cell nuclei were counterstained with DAPI (blue). The images were merged as indicated. STAT-1 is in green, nucleus is in blue, and colocalization is gray/blue.