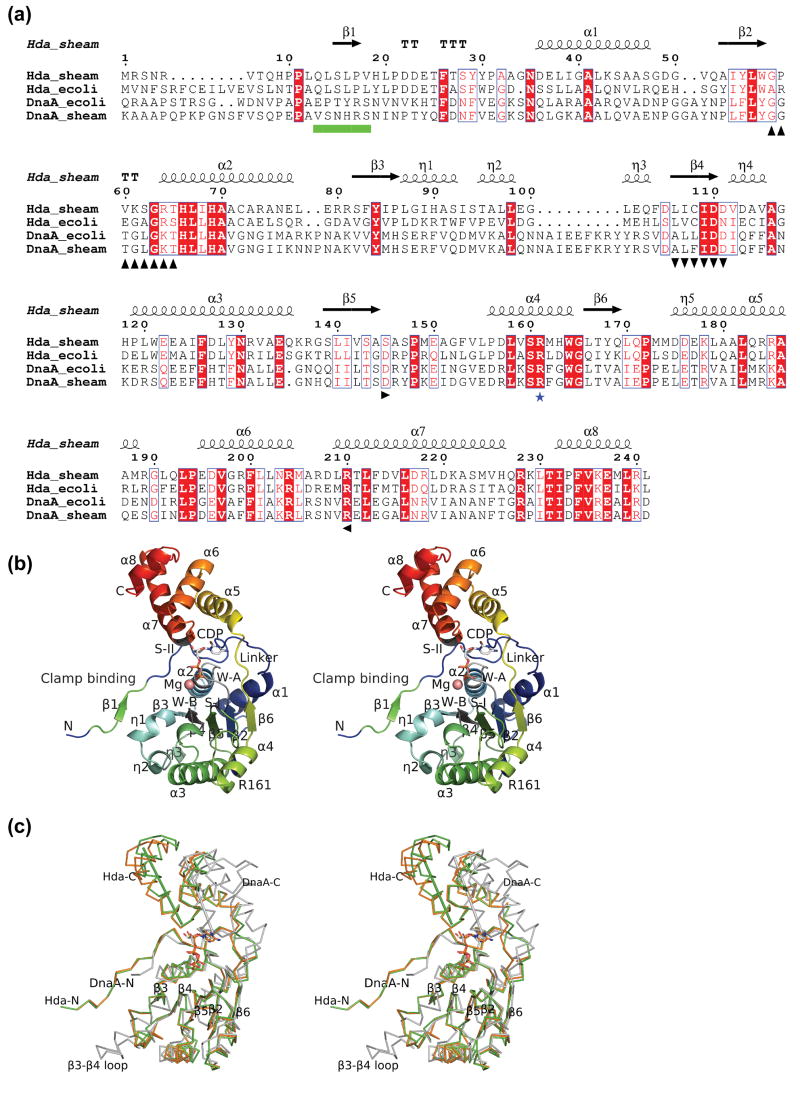
Figure 1.
Structure of Hda from Shewanella amazonensis. (a) Sequence alignment between Hda and DnaA (ATPase region) of E. coli and S. amazonensis. The Hda of S. amazonensis (Hda_sheam) is highly homologous to Hda of E. coli (Hda_ecoli) with 48% sequence identity and to the DnaA ATPase regions of E. coli (residues 107-365) and S. amazonensis (residues 95-355) with sequence identity of 24% and 21% respectively. The secondary structure elements and sequence numbering of the S. amazonensis Hda structure are shown in the top row. The conserved Walker A, Walker B, sensor I, sensor II motifs are marked in black with up, down, right and left triangles in the bottom row. The ‘arginine finger’ of box VII is shown with a blue star. The sliding clamp binding motif is denoted as green boxes. (b) Structure of an Hda monomer with bound CDP and magnesium. The Walker A (W-A, or P-loop), Walker B (W-B), sensor I (S-I), sensor II (S-II), the sliding clamp binding and box VII (‘arginine finger’) motifs are labeled corresponding to a). (c) Comparison of Hda monomers (orange and green) with the DnaA III and IIIb (PDB id 2hcb, gray). The Hda and DnaA are superimposed based on their respective NTPase domains (i.e. I and III). The sensor II helices of Hda (210–225) and DnaA (276–289) are shown as rods to highlight the significant difference in the domain Ib orientation.