Abstract
In this study we explored the relevance of Hint, a novel tumor suppressor gene, to human hepatoma. The human hepatoma cell lines Hep3B and HepG2 express very low levels of the HINT1 protein but the Huh7 cells express a relatively high level. In Hep3B and HepG2 cells, but not in Huh7 cells, the promoter region of Hint1 is partially methylated and treatment with 5-Azadcdeoxycytidine increased expression of the HINT1 protein and Hint1 mRNA in Hep3B and HepG2 cells. Increased expression of HINT1 in HepG2 cells markedly inhibited their growth. It also inhibited the transcriptional activities of β-catenin/TCF4, and USF2, and inhibited the expression of endogenous cyclin D1 and TGFβ2. Furthermore, HINT1 co-immunoprecipitated with USF2 in extracts of Hep2 cells. HINT1 also inhibited NFκB transcription factor reporter activity and inhibited translocation of the endogenous p65 protein to the nucleus of HepG2 cells. Therefore, decreased expression of the Hint1 gene through epigenetic silencing may play a role in enhancing the growth of a subset of human hepatoma by increasing the expression of genes controlled by the transcription factors β-catenin, USF2, and NFκB.
Keywords: HINT1, hepatoma, β-catenin, USF2, NFκB
Introduction
In previous studies utilizing Hint1 deficient mice we obtained evidence that this gene is a novel haplo-insufficient tumor suppressor. Thus, Hint1 deficient mice have a marked increase in susceptibility to chemical carcinogen-induced mammary tumors 1, ovarian tumors 1, and gastric tumors 2. In addition, with aging Hint1 deleted mice displayed an increase in the occurrence of a variety of spontaneous tumors, including hepatoma 1. Other investigators recently found that the Hint1 gene is transcriptionally silenced in the H522 and H538 human non-small cell lung cancer (NSCLC) cell lines and that increased expression of HINT1 inhibits the growth of these cells 3. We obtained similar results with the SW480 human colon cancer cell line 4.
There is accumulating evidence that the HINT1 protein plays an inhibitory role in several pathways that control gene transcription. Thus, in various cell systems HINT1 can interact with and inhibit the activities of cyclin-dependent kinase 7 5, the transcription factor microphthalmia (Mi) also called MITF 6, the transcription factor USF2 7, and the Pontin/Reptin/β-catenin/TCF4 complex 8. In recent studies, we obtained evidence that in SW480 colon cancer cells HINT1 interacts with the scaffold protein plenty of SH3 (POSH)-JNK2 complex, and thus inhibits the activity of the AP-1 transcription factor 4. MITF 6 and USF2 7 bind to E-box motifs in the promoter regions of several genes and thus activate their transcription. In mast cells HINT1 binds to and inhibits the transcriptional activities of both MITF and USF2 6, 7. USF2 is ubiquitously expressed in eukaryotic cells, however the expression of MITF is limited mainly to mast cells, melanocytes and osteoblasts 6, 7. USF2 plays an important role in cell proliferation by activating the expression of several genes including TGFβ2 9, 10, HIF1α 11, and PAI-1 12, 13.
The Wnt/β-catenin/TCF4 signaling pathway plays an important role in cell survival, proliferation and differentiation 8, 14. As mentioned above, in SW480 human colon cancer cells HINT1 binds to the Pontin/Reptin/β-catenin/TCF4 complex, and thus inhibits TCF4 transcription factor activity. β-catenin frequently accumulates in the nucleus of colon carcinoma cells as a result of mutations in the APC gene 14. Mutations in the APC gene have not been seen in experimental or human hepatoma 15–17. However, s-catenin is normally targeted for proteolytic degradation by phosphorylation of specific sites by GSK-3s 18, and mutations in the s-catenin gene that prevent this phosphorylation have been found in a wide variety of human carcinomas, including hepatoma 14, 17 and also in carcinogen-induced mouse hepatoma 17–20. Increased expression of s-catenin has been associated with a poor prognosis in patients with hepatoma 21.
The purpose of this study was to examine the expression and function of HINT1 in human hepatoma cells. We focused on these cells because of the above-described unusual role of β-catenin in hepatoma cells and because our Hint1 deficient mice displayed an increase in spontaneous hepatoma 1, as well as the prevalence of this disease at a worldwide level 22. Our findings suggest that Hint1 is likely to function as a tumor suppressor gene in a subset of human hepatoma.
Materials and Methods
Chemicals and reagents
5-Azadc was purchased from Sigma (St. Louis, MO), and protein G PLUS-Agarose was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell lines and cell culture
The Huh7, HepG2 and Hep3B human hepatoma cancer cell lines, the human embryonic kidney (HEK) 293 cells, and the mouse fibroblast NIH3T3 cell line were obtained from American Type Culture Collection (Manassas, VA). The PT67 retrovirus packaging cell line, originally derived from human embryonic kidney cells, was from CLONTECH Laboratories, Inc (Palo Alto, CA). All of the cell lines were maintained in DF10 medium containing DMEM (Invitrogen, San Diego, CA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), and were incubated in a 100% humidified incubator at 37°C with 5% CO2. Colony formation and cell proliferation assays were done as previously described4.
Methylation specific PCR
Whole cell DNA was isolated from cultured cells, modified with bisulfite and subjected to PCR amplification using PCR primer sets for detection of methylated CpG dinucleotides within a CpG island in the promoter region of the Hint1 gene, as previously described4.
5–Azadc Treatment and Semi-quantitative RT-PCR
Cells were treated with solvent or with 5-Azadc (1μM) for 96 hours. Total RNA was extracted with a single step method, RT-PCR was conducted using Hint1 primers and the products analyzed on a 1% agarose gel as previously described4.
Plasmid construction, gene transduction and proliferation assays
We have previously described the construction of the HA-Hint1 wild type (WT) and HA-Hint1 His112/Asn112 mutant plasmids; the retroviral packaging system and the retrovirus mediated Hint1 transduction procedure; cell proliferation assays; and colony formation assays4.
Protein extraction and western blot assays
Cells were cultured and treated with the various conditions as described above. The whole cell protein extracts were then prepared, as described previously 4. The specific cytoplasmic protein fraction and the nuclear protein fraction were prepared using protein separating kits (Nuclear Extract Kit, Active Motif, Carlsbad, CA) according to the manufacturer’s instructions. The proteins were separated by SDS-PAGE with 7.5 to 12.5% polyacrylamide gels and then blotted with the indicated antibody. The primary antibodies included HINT1 antiserum 2, anti-HA-Tag antibody (Sigma), anti-cyclin D1 antibody (Upstate Biotechnology Inc., Lake Placid, NY), anti-β-catenin antibody (Santa Cruz), anti-USF2 antibody(Santa Cruz), anti-IκBα antibody (Cell signal), anti-NFκBp65 antibody (Chemicon, Tenecula CA), anti-β-tubulin antibody (Cell signal) and anti-β-actin antibody (Sigma). Anti-mouse and anti-rabbit IgG (Amersham Biosciences) antibodies were used as the secondary antibodies. Each membrane was developed with an ECL enhanced chemiluminescence system (Amersham Biosciences). The intensities of specific protein bands were quantified with NIH Image software version 1.62. corrected for the intensity of the respective β-actin band, or β-tubulin band, and expressed as a ratio with respect to a control lane.
Luciferase reporter assays
Cells were plated at 1 × 105 cells per 35-mm-diameter plates 18 hours before transfection. The cells were transfected with 400 ng/well of the indicated luciferase reporter plasmid which in specific experiments included: Topflash (three copies of the optimal Tcf motif) Fopflash (three copies of a mutant Tcf motif), p1745CD1 LUC (which contains intact SP1, TCF, CRE and NF-κB sites in the full length 1745 bp cyclin D1 promoter), pNF-κB -Luc, E-box-LUC (kindly provided by Kathryn Calame, Columbia University), cathepsin K-LUC, and 200 ng of the pCMV-β-galactosidase reporter plasmid, with or without co-transfection with other indicated plasmid DNAs (see Figure legends). The latter included: β-catenin WT, pHA-Hint1-WT and USF2 WT. All transfections were done with the Lipofectin reagent (Invitrogen, Carlsbad, CA). At 36 hours post transfection, cell extracts were prepared, and each sample was assayed, in triplicate, using a luciferase assay system (Promega Corp., Madison, WI). Luciferase activities were normalized to β-galactosidase activities to correct for differences in transfection efficiency and expressed as Relative (Rel) Luciferase (LUC) activity. Significant differences (p < 0.05) with respect to the corresponding control assay are indicated by an asterisk (*).
TGFβ2 CAT ELISA assays
The p77 (E-box wide type), p77E (E-box mutant) and p40 (mutant without E-box) TGFβ2 CAT reporter plasmids were kindly provided by Angie Rizzino (University of Nebraska Medical Center, Omaha, Nebraska). The TGFβ2 CAT reporter plasmids (0.5 μg), the pCMV-β-galactosidase reporter plasmid (0.2 μg), with or without co-transfection with the indicated plasmid DNAs (see Figure legends), were transiently transfected into the HepG2 cells, as described above. After 36h cell extracts were prepared and CAT activity was measured by using an ELISA kit (CAT ELISA; Roche Diagnostics Co.) according to the manufacturer’s instructions. The level of CAT activity found in cells transfected with the empty p40-CAT plasmid was set to a value of 1, and changes observed in the treated cultures were expressed as relative CAT activity.
Immunoprecipitation
Cells were plated into 10-cm-diameter plates 18 hours before transfection. Then the cells were transfected with pCMV-USF2 plasmid DNA (2.5 μg) 23, and co-transfected with pHA-Hint1-WT (2.5 μg), plasmid DNA, alone or together, as indicated. After 48 hours cells were washed twice with the cold PBS, and incubated with ice-cold lysis buffer (10 mM Tris pH 7.4, 1.0% Triton X-100, 0.5% NP-40, 150mM NaCl, 20mM NaF, 0.2mM sodium orthovanadate, 1 mM EDTA, 1 mMEGTA, 0.2 mM PMSF) for 15 minutes at 4°C. Immunoprecipitation of specific protein complexes in these cell lysates (200 μg protein) was performed with 4 μg of an anti-HA M2 affinity gel (Sigma) which was pre-bound to Protein-A agarose, or an anti-β-catenin monoclonal antibody or an anti-USF2 polyclonal antibody together with protein-G agarose (Sigma), for 18 h at 4°C. The immunocomplexes were washed five times with cold cell lysis buffer and then boiled for 5 minutes. The samples were then analyzed by Western blots using the indicated antibodies.
Reproducibility and statistical analysis
The Western blots and the RT-PCR results are representative of duplicate or triplicate assays that gave similar results. The colony formation, growth curve, and reporter assays were done in triplicate. These data are plotted as means and S.D.s and were analyzed using Student’s t test. A difference between groups with p < 0.05 was considered statistically significant.
Results
Expression of Hint1 in Hepatoma cells
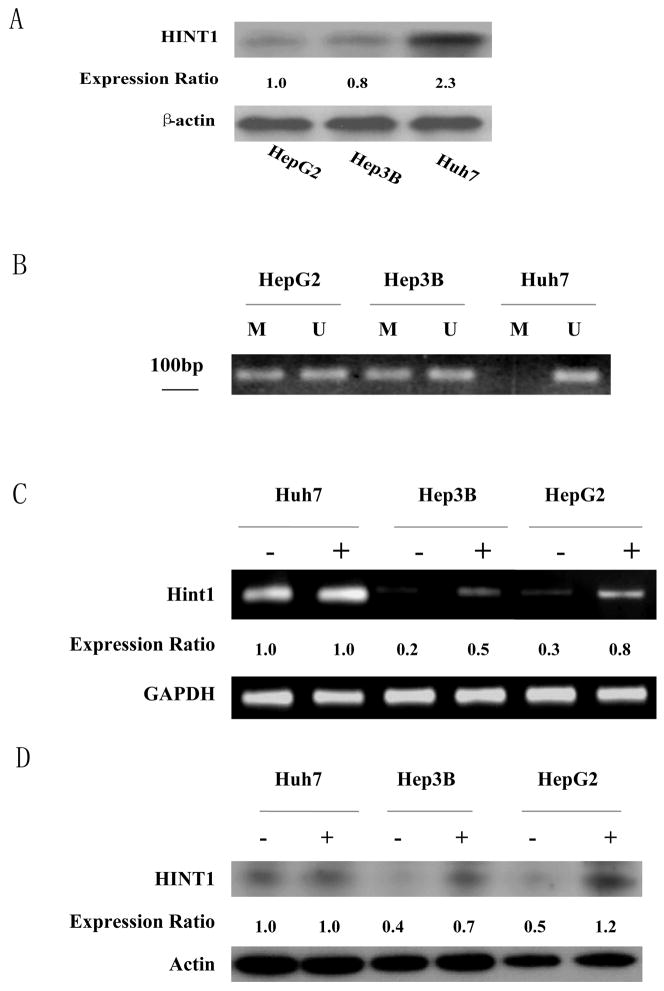
Western blot analysis indicated that amongst the 3 hepatoma cancer cell lines examined HepG2 and Hep3B cells displayed a low level of HINT1, but the Huh7 cells expressed about twice this level of HINT1 (Fig. 1A). These results were reproduced in additional studies. Therefore, we examined the state of methylation of a CpG rich region in the promoter region of the Hint1 gene using bisulfite treated DNA samples obtained from the above 3 cell lines, employing sets of methylation specific primers, as described in Materials and Methods. No methylation of this region of DNA was detected in the DNA sample from the Huh7 cells (Fig. 1B), which is consistent with the relatively high level of expression of the HINT1 protein in these cells (Fig. 1A). However, both methylated and unmethylated bands were detected in the HepG2 and Hep3B DNA samples (Fig. 1B).
Figure 1. Levels of expression of the endogenous HINT1 protein and extent of methylation of the Hint1 promoter in human hepatoma cell lines.
(A), Western blot analysis of HINT1 expression in extracts of the indicated cell lines. Actin was used as an internal loading control. HINT1 protein levels were quantified by densitometry and normalized to the corresponding level of actin. The expression ratios between the cell lines are presented in the lower panel. (B), Methylation Specific PCR (MSP). Total cellular DNA was extracted from the indicated cell lines, modified with bisulfite, and subjected to PCR with sets of specific PCR primers (U, unmethylated; M, methylated). PCR products were analyzed on 1% agarose gels and visualized under UV illumination. All assays ware repeated three times, and gave similar results. (C), Cells were treated with 5-Azadc (1μM) for 96 hours. Total RNA was isolated and semi-quantitative RT-PCR was done with specific Hint1 and GAPDH (internal loading control) primers. PCR products were analyzed on 1% agarose gels and visualized under UV illumination. (−) no treatment, (+) 5-Azadc. Expression ratios for Hint1 mRNA were calculated after normalization for GAPDH. All assays were repeated three times and gave similar results. (D), Cells were treated with 5-Azadc as described above, and HINT1 expression levels were detected by Western blot analysis. Actin was used as loading control. (−) no treatment, (+) 5-Azadc. Expression ratios for HINT1 protein were calculated after normalization for β-actin. Assays were done in duplicate and gave similar results.
These results suggested that the relatively low levels of HINT1 in the HepG2 and Hep3B cells may be due, at lease in part, to promoter methylation. Therefore, we treated the above 3 cell lines with a 1μM concentration of the DNA demethylating agent 5-azadeoxycitidine (5-Azadc) for 96 hrs and then examined by RT-PCR and western blots the levels of expression of HINT1 mRNA and protein, respectively. In both the HepG2 and Hep3B cells treatment with 5-Azadc led to about a 2 fold increase in expression of Hint1 mRNA (Fig. 1C) and protein (Fig. 1D). In contrast, similar treatment of Huh7 cells with 5-Azadc did not increase the levels of Hint1 mRNA or protein (Figs. 1C and 1D), which is consistent with our evidence that the Hint1 promoter is not methylated in Huh7 cells (Fig. 1B). Taken together, the studies described in Fig. 1 provide evidence that the relatively low level of expression of Hint1 in HepG2 and Hep3B cells is due, at least in part, to partial methylation of the promoter region of this gene.
Increased expression of HINT1 inhibits growth of HepG2 cells
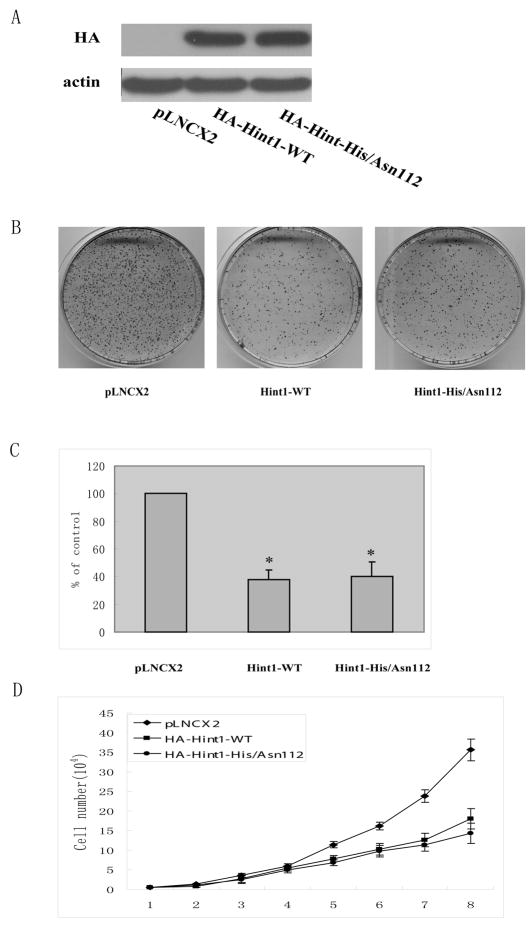
We utilized retrovirus expression vectors that encode either the HA-tagged wild type Hint1 cDNA sequence (Hint1-WT) or a HA-tagged mutant Hint1 sequence in which His residue 112 was replaced with Asp (Hint1/His/Asp112). This mutant is of interest since this substitution might disrupt the HIT motif that is characteristic of HINT1 and related members of the HIT superfamily of proteins 1, 2. Infection of HepG2 cells with either of these vectors caused approximately similar overexpression of the related HA-tagged proteins (Fig. 2A). The infected cells were then analyzed for their colony forming efficiency. We found that the overexpression of either WT-Hint1 or Hint1-His/Asp 112 caused about 61% inhibition of colony formation (p<0.05) when compared to the control pLNCX2 vector infected cells (Figs 2B, C). We also carried out cell proliferation assays over a period of 8 days and found that both the Hint1-WT and Hint1-His/Asp112 infected cells displayed about a 50% inhibition of cell proliferation when compared to the vector control infected cells (Fig. 2D).
Figure 2. Overexpression of HINT1 inhibits growth of HepG2 cells.
The retrovirus shuttle plasmid pLNCX2 encoding HA-Hint1-WT, or HA-Hint1-His/Asn 112, or an empty control plasmid, was transfected into the PT67 packaging cell line. After incubation for 48h the retrovirus containing medium was harvested. HepG2 cells were then infected with the indicated retroviruses for 24 h at a multiplicity of infection of 800 and then incubated in fresh medium for 72 h. (A), Total cell lysates were prepared and Western blots performed with an Anti-HA antibody. (B) and (C), Colony formation assay. The infected cells were grown in the presence of the selection agent G418 for 3 weeks. The colonies were then fixed, stained with Giemsa solution, and counted. The photographs (B) display representative plates and the bar graph (C) displays the percent inhibition of colony formation. The results are the means and standard deviations (S.D.) of triplicate assays. The asterisk * indicates significant inhibition (p<0.01). (D) Growth curves. Cells were cultured in 24 well plates, infected with the above described retrovirus-containing medium and the number of cells in replicate wells was counted every day for the subsequent 8 days. The data indicate the mean values and S.D.s of triplicate assays. Similar results were obtained in a repeat assay.
HINT1 inhibits β-catenin/TC4 activity and cyclin D1 expression in HepG2 cells
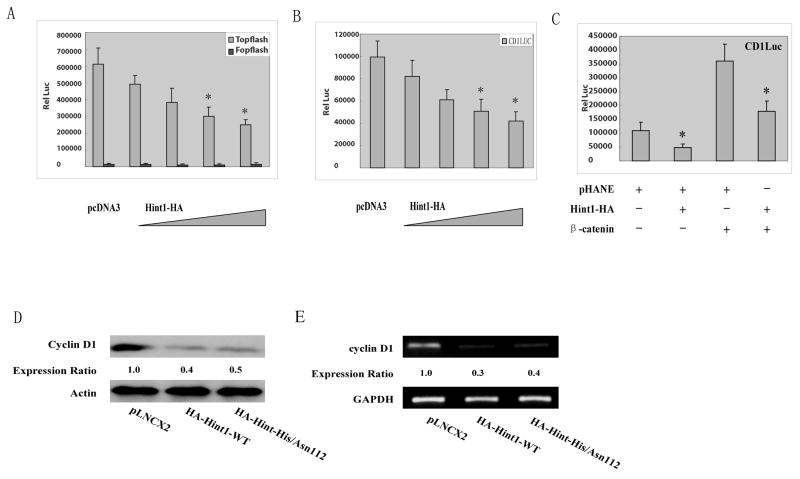
A previous study in APC mutant SW480 human colon cancer cells demonstrated that HINT1 binds to the proteins Pontin and Reptin, which complex with β-catenin, and that this inhibits the β-catenin/TCF4 signaling pathway in these cells 8. However, in contrast to SW480 cells HepG2 cells have wild type APC, but instead express a truncated β-catenin protein 24. In addition, in a previous study we found that overexpression of wild type β-catenin causes marked activation of the transcription factor activity of the cyclin D1 promoter in HepG2 cells 24. Therefore, it was of interest to examine the effects of HINT1 on β-catenin/TC4 activity and on cyclin D1 promoter activity in HepG2 cells since the β-catenin pathway is altered in a different manner than in SW480 cells. When HepG2 cells were transfected with the “Topflash” luciferase reporter which contains three copies of the optimal Tcf motif they displayed high luciferase activity, presumably reflecting the high activity of the truncated endogenous β-catenin protein. No activity was obtained with the negative control Fopflash reporter (Fig. 3A). Co-transfection with increasing concentrations of pHA-Hint1-WT plasmid DNA produced a concentration-dependent inhibition of this activity; two μg of this DNA caused about 60% inhibition (Fig. 3A). Therefore, it appears that HINT1 can inhibit transcriptional activity of both the wild type β-catenin 9 and a truncated β-catenin protein.
Figure 3. HINT1 inhibits β-catenin/TCF4 transcription factor activity and cyclin D1 expression in HepG2 cells.
(A) The Topflash luciferase reporter and CMV-β-gal reporter plasmid DNAs were transfected into the HepG2 cells. As indicated, increasing amounts of pHA-Hint1 plasmid DNA (0, 0.5, 1.0, 1.5, 2.0/well) were also co-transfected, and pcDNA was added, as needed, to achieve the same total amount of plasmid DNA in each assay. Luciferase activity was assayed at 36 h post-transfection and normalized for β-gal activity. All assays were done in triplicate and the S.D.s are shown. Similar results were obtained in a repeat study. No significant luciferase activity was obtained with the negative control Fopflash luciferase reporter. (B) The cyclin D1 reporter CD1 (-1745)-LUC and CMV-β-gal reporter plasmid DNAs were co-transfected into HepG2 cells, with our without increasing amounts of Hint1 plasmid DNA and luciferase activity assayed 36 hours post transfection as described in Fig. 3A. (C) the cyclin D1 reporter assays were done as in (B), with co-transfected Hint1-HA or wild type β-catenin plasmid DNAs, as indicated. (D) HepG cells were infected with the indicated retrovirus containing medium as described in Fig. 2A. At 72 hours post infection, whole cell protein extracts were prepared and the levels of cyclin D1 protein expression were determined by Western blot analysis. Actin was used as loading control. Expression ratios for cyclin D1 protein were calculated after normalization for β-actin. (E) HepG2 cells were treated as in (D) and at 36 hours cellular RNA was extracted and semi-quantitative RT-PCR was performed using specific primer sets for cyclin D1 and GAPDH. RT-PCR products were analyzed on 1% agarose gels and visualized under UV illumination. Expression ratios for cyclin D1 mRNA were calculated after normalization for GAPDH. All assays were repeated twice and gave similar results.
One of the major effects of the β-catenin/TCF4 signaling pathway is stimulation of transcription of the cell cycle control protein cyclin D1 22. When a full length cyclin D1 promoter-luciferase reporter (-1745CD1LUC) was transfected into HepG2 cells co-transfection with increasing amounts of Hint1 plasmid DNA caused a concentration dependent inhibition of the transcriptional activity of the cyclin D1 promoter (Fig. 3B). Transfection with plasmid DNA encoding wild type β-catenin markedly stimulated this reporter activity in HepG2 cells and additional co-transfection with Hint1 plasmid DNA largely inhibited this stimulatory activity (Fig. 3C). To directly examine the effects of Hint1 on expression of the endogenous cyclin D1 gene in HepG2 cells these cells were infected with the retrovirus vector encoding the Hint1 cDNA sequence, as described in Fig.2A. We found that overexpression of either WT-Hint1 or Hint1-His/Asn112 inhibited the expression of both endogenous cyclin D1 protein (Fig. 3D) and cyclin D1 mRNA(Fig. 3E) in HepG2 cells.
Taken together, the above data indicated that HINT1 inhibits the transcriptional activities of both the truncated β-catenin protein present in HepG2 cells (Fig. 3A) and also an exogenous wild type β-catenin protein when it is expressed in these cells (Fig. 3C). These effects probably explain why we also found that Hint1 inhibits expression of endogenous cyclin D1 in HepG2 cells (Fig. 3D), although other effects of HINT1 on gene expression may also contribute to the latter effect.
HINT1 inhibits USF2/E-box related transcription factor activity and TGFβ2 expression in HepG2 cells
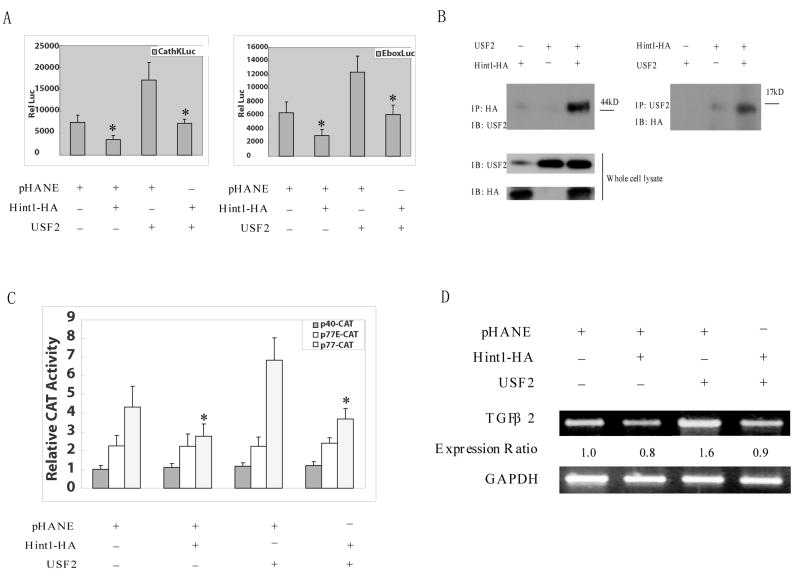
Previous studies indicated that in mast cells HINT1 binds to and inhibits the transcriptional activities of the bHLH transcription factors MITF and USF2 6, 7. The expression of MITF is restricted to cells in the neuroectodermal lineage 6 but USF2 is expressed in a variety of tissues 7. Indeed, we found that USF2 but not MITF is expressed in HepG2 cells and other human cancer cell lines (data not shown). Therefore, it was of interest to determine if HINT1 inhibits the transcriptional activity of USF2 in HepG2 cells. Since USF2 binds to the E-box motif in the promoter region of target genes 7, 9 we did transient transfection reporter assays employing a cathepsin K-LUC reporter which contains an E-box element. In this assay HepG cells had an appreciable basal level of activity and this was inhibited about 50% by co-transfection with the Hint1 plasmid DNA. Overexpression of USF2 stimulated cathepsin K-LUC activity and this stimulation by USF2 was abrogated by cotransfection with the Hint1 plasmid DNA (Fig. 4A, left panel). Very similar inhibitory effects of Hint1 on USF2 activity were obtained in HepG2 cells with a p-E-box-LUC reporter that contains two repeat E-box elements (Fig. 4A, right panel).
Figure 4. HINT1 inhibits USF2/E-box related transcription factor activity and TGFβ2 expression in HepG2 cells.
(A), The p-cathepsin K-LUC or pE-box-LUC, and CMV-β-gal reporter plasmid DNAs, were co-transfected into HepG2 cells. pHANE, pHA-Hint1 and pUSF2 plasmid DNAs were also co-transfected, as indicated. Luciferase activity was assayed 36 hours later and corrected for β-gal activity, as described in Fig. 3A. All assays were done in triplicate and a repeat study gave similar results. (B), HepG2 cells were transfected with pCMV-USF2 or pHA-Hint1 plasmid DNAs as indicated. After 48 hours, whole cell protein extracts were prepared and were immunoprecipitated with an anti-HA monoclonal affinity gel (left panel) or an anti-USF2 polyclonal antibody together with protein G plus-agarose (right panel), at 4°C for 18 hours. The immunoprecipitates were washed with cold lysis buffer five times, and then analyzed on Western blots with an anti-USF2 polyclonal antibody or an anti-HA monoclonal antibody to detect the USF2 or the HA tagged HINT1 proteins in the immunocomplexes and in the whole cell lysate (bottom panel), as indicated. (C), HepG2 cells were transfected with wild type (p77-CAT) or mutant (p40-CAT, p77E-CAT) TGF-β2 promoter/CAT reporter plasmid DNAs together with the CMV-β-gal reporter plasmids DNA. At 36 hour post transfection, whole cell extracts were prepared, and CAT activity was assayed and normalized for β-gal activity. The results are expressed as the CAT activities of the pβ2-77 or pβ2-77E relative to the activity of the pβ2-40 construct. All assays were done in triplicate and a repeat study gave similar results. (D), pHA-Hint1 or the pCMV-USF2 plasmid DNAs were transfected into HepG2 cells, as indicated. At 48 hours post transfection, whole cell RNA was extracted, and semi-quantitative RT-PCR was performed using specific TGF β2 and GAPDH primer sets, as described in Fig. 4D. Assays were repeated twice and gave similar results.
In view of the above results it was of interest to determine whether HINT1 binds to USF2 in intact HepG2 cells. Therefore, HepG2 cells were transfected with expression vectors encoding USF2 and HA-tagged HINT1, either alone or in combination, and cell extracts prepared. When proteins in these extracts were immunoprecipitated with an anti-USF2 polyclonal antibody and analyzed by western blots using an anti-HA monoclonal antibody we observed a distinct HINT1 protein band only in the extract from cells transfected with both the USF2 and Hint1-HA plasmids (Fig. 4B, right panel, lane 3). In a reciprocal experiment when the cells extracts were immunoprecipitated with an anti-HA antibody and the western blots probed with an anti-USF2 antibody we observed the USF2 protein in the extract of cells that had been co-transfected with both the Hint1 and USF2 plasmids (Fig. 4B, left panel, lane 3). Similar co-immunoprecipitation results were obtained when the same studies were done with HEK 293 cells (data not shown).
Several target genes for USF2 have been identified in cancer cells that play a role in tumorigenesis, including the TGF-β2 gene 9, 10. Indeed the promoter of the TGF-β2 gene has an E-box site that is localized in a critical positive regulatory region with respect to transcription 9, 10. Therefore, we examined the possible effects of USF2 and HINT1 on the transcriptional activity of a TGF-β2 promoter construct, and also the effects of HINT1 on expression of the endogenous TGF-β2 gene in HepG2 cells. In the reporter assays we used a series of three constructs. p77-CAT, a wild type TGF-β2 derived promoter that contains both an E-Box and a CRE/ATF element linked to CAT; p77E-CAT, the same construct but with a base substitution in the E-Box element; and p40-CAT, a construct in which both the E-Box and the CRE/ATF elements are missing (see “Materials and Methods”). The negative control reporter p40-CAT gave low activity and the mutant E box reporter p77E-CAT gave somewhat higher activity in HepG2 cells but neither of their activities was significantly influenced by co-transfection with Hint1 or USF2 (Fig. 4C). The wild type p770-CAT reporter gave relatively higher activity and this activity was significantly inhibited by co-transfection with the Hint1 plasmid and stimulated by co-transfection with the USF2 plasmid. The latter simulation was abrogated when the cells were also co-transfected with Hint1 (Fig. 4C).
To obtain more direct evidence that USF2 and HINT1 affect the expression of TGF-β2, HepG2 cells were tansfected with pHA-Hint1 or pUSF2 plasmid DNAs, either separately or in combination. Forty-eight hours later total mRNA was extracted and semi-quantitative RT-PCR was preformed for TGF-β2 mRNA. Transfection with Hint1 caused about a 20% decrease, whereas co-transfection with USF-2 caused an about 1.6 fold increase, in the basal level of TGF-β2 mRNA. Co-transfection with Hint1 abrogated the stimulation of TGF-β2 mRNA expression caused by USF2 (Fig. 4D). Similar results were obtained in a repeat experiment (data not shown).
HINT1 inhibits NFκB transcription factor activity in HepG2 cells
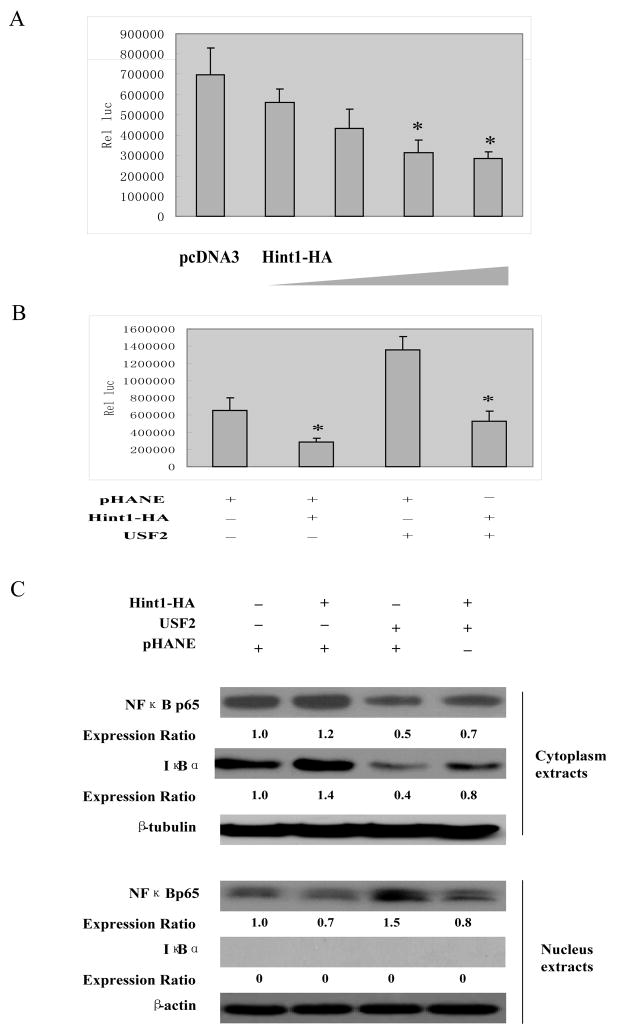
In their inactive state heterodimers of p50 and p65, also known as classical NF-κB, are retained in the cytoplasm and are bound to the IκB family of scaffold proteins. This blocks nuclear translocation and the transcriptional activation of NFκB. The mechanism of activation of NFκB is not entirely clear but it appears to be due to phosphorylation and subsequent ubiquitin/proteosome mediated degradation of the IκB protein thus allowing for nuclear translocation of NFκB and increased expression of NFκB responsive genes 25. Previous studies have shown that in some cancer cell lines TGF-β can enhance NFκB activation and thereby increase cell survival, although the precise mechanism is not known 26. In view of the above described effects of USF2 and HINT1 on TGFβ2 expression (Fig. 4), we examined their effects on NFκB activation in HepG2 cells. When HepG2 cells were transfected with a pNFκB -LUC reporter plasmid DNA, they displayed high luciferase activity (Fig. 5A). Co-transfection with increasing concentrations of a pHA-Hint1-WT plasmid DNA produced a concentration-dependent inhibition of this activity. Controls included an empty pHANE plasmid and all assays were normalized for CMV-β-gal reporter activity. We also found that transfection with USF2 plasmid DNA stimulated NFκB activity while co-transfection with Hint1 plasmid DNA largely abrogated this stimulation by USF2 (Fig. 5B).
Figure 5. HINT1 inhibits NF-κB transcription factor activity in HepG2 cells.
(A) and (B), pNF-κB-Luc (400 ng/well) and the CMV-β-gal reporter control plasmid DNAs, together with pHA-Hint1 (0, 0.5, 1.0, 1.5, 2.0 μg) in (B) or pHA-Hint1 (1.0 μg) with our without CMV-USF2 (1.0 μg) plasmid DNAs, as indicated, were transfected into the HepG2 cells. Thirty-six hours later luciferase activity was assayed as described in Fig. 3A. (C), pHA-Hint1 or the CMV-USF2 plasmid DNAs were transfected into HepG2 cells, as indicated. At 48 hours post transfection, cytoplasmic and nuclear proteins were extracted, and each fraction was analyzed by Western blots to detect the expression levels of p65, IκBα, β-tubulin and β-actin. The latter two proteins were used as loading controls for the cytoplasmic and nuclear extracts, respectively. Expression ratios were calculated after normalization for β-tubulin and β-actin. Assays were repeated three times and gave similar results.
As described above, activation of NFκB transcription factor activity is due to degradation of the IκB protein and nuclear translocation of the NFκB protein. Therefore, we examined possible effects of USF2 and HINT1 on cytoplasmic and nuclear levels of the NFκB 65 and IκBα proteins. HepG2 cells were transfected with pHA-Hint1 or USF2 plasmid DNAs, either separately or in combination, and 48 hours post transfection, specific nuclear and cytoplasmic protein extracts were prepared and each extract was examined in parallel by Western blot analyses with a p65 antibody and an IκBα antibody (Fig. 5C). β-tubulin was used as a protein loading control for the cytoplasmic extract and β-actin as a control for the nuclear extract. We found that with the extracts from the untreated HepG2 cells the NFκBp65 protein was detected in both the cytoplasmic and nuclear extract indicating constitutive activation of this transcription factor in these cells. As expected, the IκBα protein was found in the cytoplasmic extract but not the nuclear extract of the control and treated HepG2 cells. Therefore, the nuclear extract was not significantly contaminated with cytoplasmic proteins. In the Hint1 transfected cell there was an increase in the intensity of the cytoplasmic NFκBp65 and IκBα bands and a decrease in the intensity of the nuclear NFκBp65 band. This finding is consistent with the evidence presented in Fig. 5A that HINT1 inhibits the transcriptional activation of NFκB. On the other hand, in the USF2 transfected cells (Fig. 5C, lane 3), there was a decrease in the intensity of the NFκBp65 protein band in the cytoplasmic extract and an increase in the intensity of this band in the nuclear extract; and these changes were associated with a decrease in the IκBα band in the cytoplasmic extract. The results shown in lane 4 of Fig. 5C indicate that co-transfection of the cells with USF2 plus Hint1 partially abrogated the effects of USF2 on nuclear translocation of NFκBp65 and NFκB degradation shown in lane 3 of Fig. 5C. Similar results were obtained in a repeat experiment (data not shown). These findings are consistent with the evidence in Fig. 5B that USF2 stimulates the transcriptional activity of NFκB and that Hint1 can abrogate this effect. Taken together, the results shown in Figs. 5A, B and C suggest that USF2 stimulates the transcriptional activity of NFκB by enhancing the degradation of IκBα. We suggest that these effects of USF2 and HINT1 on IκBα stability and NFκB activation are mediated at least in part via TGFβ in view of the results we obtained in Figs. 4C and D and previous studies implicating TGFβ in NFκB activation in cancer cells 26. However, the precise mechanism by which TGFβ exerts this effect is not known.
Discussion
In previous studies we found that mice with deletions in the Hint1 gene are more prone to develop spontaneous hepatoma 1. In view of this finding and the prevalence of hepatocellular cancer in Asia and Africa and the rising incidence and mortality of this disease in the United States 22, in the present study we examined several molecular aspects of the function of HINT1 in human hepatoma cells. We found a low level of expression of the HINT1 protein in the HepG2 and Hep3B human hepatoma cell lines when compared to the Huh7 human hepatoma cancer cell line, and obtained evidence that this is due to methylation of the promoter region of the Hint1 gene in HepG2 and Hep3B cells (Fig. 1). Similar findings have been obtained in subsets of human lung cancer 3 and colon cancer cell lines 4. Furthermore, we found that increased expression of HINT1 inhibited the growth of HepG2 cells (Fig. 2) and Hep38 cells (data not shown).
Although previous studies 8 indicated that in SW480 colon cancer cells HINT1 inhibits the transcriptional activity of β-catenin/TCF4 by binding to the associated Pontin/Reptin complex, the present studies provide the first evidence that HINT1 also inhibits β-catenin/TCF4 activity in hepatoma cells (Fig. 3). This is of interest because in SW480 cells, and colon cancer cells in general, β-catenin is activated because of mutations in APC 8, 14 whereas in hepatocarcinomas it is activated by other mechanisms, including mutations in β-catenin 15–20. Furthermore, about 50–70% of clinical samples of hepatocellular carcinoma display an accumulation of β-catenin 27. A recent study indicates that the FHIT protein, a known tumor suppressor, which like HINT1 is a member of the HIT family of proteins, also inhibits β-catenin activity but does so by binding directly to the C-terminus of β-catenin rather than to pontin or reptin 28. To our knowledge this has not been examined in hepatoma cells. In mast cells HINT1 inhibits the activities of the bHLH transcription factors MITF and USF2 by binding directly to these proteins 6, 7. Unlike MITF which is only expressed in neuroectoderm-derived cells 6, USF2 is ubiquisiouly expressed in epithelial cells 7. Both USF1 and USF2 transcription factors play important roles in regulating various metabolic processes in hepatoma cells 29, 30 However, whether USF1 or USF2 also play a role in the proliferation of these cells has not been studied. We found that USF2 is expressed at a relatively high level in HepG2 cells (data now shown). During transcription USF2 binds to an E-box motif in the promoter region of several genes 7. Indeed we found that in HepG2 cells transfection with a USF2 plasmid DNA stimulated the transcriptional activity of a cathepsin K promoter which contains an E-box element, while co-transfection with a Hint1 plasmid largely abrogated this stimulation by USF2 (Fig. 4A). Similar results were obtained when these assays were performed with a p-E-box LUC reporter that contains two repeat E-box elements (Fig. 4A). Co-immunoprecipitation studies indicated that HINT1 binds to the USF2 protein in HepG2 cells (Fig. 4B) and similar results were obtained in HEK293 cells (data not shown). Thus, HINT1 can inhibit USF2 transcription factor activity in hepatoma cells by directly binding to USF2. Although this had been previously shown in mast cells 7 this is the first evidence that this occurs in human carcinoma cells. As reviewed in the Introduction, the TGF-β2 gene is a specific target of USF2. We found that transfection of HepG2 cells with USF2 markedly increased the transcriptional activity of an E-box containing sequence present in the TGF-β2 promoter, and that co-transfection with a Hint1 plasmid inhibited this activation (Fig. 4C). Furthermore, increased expression of USF2 also induced increased expression of TGF-β2 mRNA in HepG2 cells and co-transfection with a Hint1 plasmid inhibited this activation (Fig. 4D). Thus far, we have not been able to detect an increase in TGF-β2 protein in the cells overexpressing USF2 (data not shown), but this may reflect lack of sensitivity of our antibody.
Although USF2 is often considered a transcription factor that inhibits cell proliferation, it can also positively regulate several genes that stimulate cell proliferation, including cyclin B1 31, CDK1 32 and CDK4 33. Furthermore, homologous deletion of USF2 causes reduced body size in mice, both at birth and throughout postnatal development 34. Therefore, in some cell types USF2 may exert a positive effect on cellular proliferation. TGF-β2 is another bi-functional regulator of cellular growth and differentiation, whose effects are highly cell type specific 35, 36. Secreted TGF-β2 activates NFkB which enhances cell survival, and suppression of TGF-β2 is growth inhibitory in PC3 prostate cancer cells 26. In addition, increased expression of TGFβ2 is associated with advanced stage in gastric tumors and this correlates with a poorer prognosis 37.
The NFκB transcription factor plays an important role in tumor development 25. A previous study demonstrated that TGFβ2 inhibits the activation of NFκB, although the mechanism is not known 26. In the present study reporter assays indicated that, as expected, there is high NFκB transcription factor activity in HepG2 cells. However, co-transfection with increasing concentrations of a Hint1 plasmid DNA produced a concentration-dependent inhibition of this activity (Fig. 5A). We also found that in HepG2 cells transfection with a USF2 plasmid stimulated NFκB activity, while co-transfection with a Hint1 plasmid largely abrogated this stimulation by USF2 (Fig. 5B). Activation of NF-κB transcription factor activity is due to degradation of IκB and nuclear translocation of NF-κB 25. Western blot analysis of nuclear and cytoplasmic extracts prepared from untreated HepG2 cells revealed that the NF-κB p65 protein was present in both the nucleus and the cytoplasm, providing evidence for its constitutive activation (Fig. 5C, lane 1). Transfection of HepG2 cells with the USF2 plasmid DNA decreased the cytoplasmic levels of both the p65 and IκB proteins, and increased the nuclear level of the p65 protein (Fig. 5C, lane 3), providing direct evidence that USF2 induced activation of NF-κB. However, transfection of HepG2 cells with Hint1 maintained IκB stability, and blocked nuclear translocation of p65. Similar effects of Hint1 were seen in HepG2 cells co-transfected with USF2 (Fig. 5C, lane 2, lane 4). These data are consistent with results obtained in the above described NF-κB reporter assays (Fig. 5A). Taken together, these results provide evidence that HINT1 inhibits NF-κB transcription factor activity by maintaining the stability of the cytoplasmic scaffold protein IκB and thereby blocking nuclear translocation of p65. Several studies indicate that the TGFβ family of proteins activate the NF-κB pathway by causing phosphorylation of IκB and IκB degradation 38–40. Thus, NF-κB can be activated in cancer cells through secretion of cytokines such as TGFβ2 41. Since USF2 activates TGFβ2 gene transcription in HepG2 cells (Fig. 4D), the inhibition by HINT1 of NF-κB nuclear translocation and NF-κB transcriptional activity may be due to the transcriptional activity of USF2, thus decreasing TGFβ2 and stabilizing IκB. Our discovery that HINT1 can inhibit Ap-1 activity 4 may also be relevant to the ability of HINT1 to inhibit TGF-β signaling since it was recently reported that AP-1 is an essential cofactor for TGF-β-dependent transcription of many genes 42.
In contrast to our finding of an interaction between HINT1 and USF2 in hepatoma cells (Fig. 4B), we did not observe any interaction of HINT1 with USF1 in these cells, either under conditions similar to those used for co-immunoprecipitation studies with USF2, or even less stringent conditions (data not shown). This is consistent with the previous findings in mast cells, in which HINT1 co-precipitated specifically with USF2 but not USF1 7. Because USF1 and USF2 frequently form heterodimers 43, it was of interest that HINT1 interacts with USF2, but not USF1, in both mast cells 7 and hepatoma cells (Fig. 4B and unpublished studies). Since USF2 can also form a homodimer 43–45, it is possible that HINT1 associates with the homodimer but not the USF1/USF2 heterodimer. It was previously reported that USF1/USF2 heterodimers and USF2 homodimers have opposite roles in the transcription of ribosomal RNA 46. Although it is not known whether USF homodimer and heterodimer formation provide a mechanism for regulating the activation of other USF target genes, this possibility is supported by the finding that BRCA1 associates with USF2 but not USF1 in co-regulating the expression of BRCA1 target genes 47.
In summary, the present study indicates that HINT1 inhibits the activities of the β-catenin/TCF4, USF2 and NF-κB transcription factors in HepG2 human hepatoma cells. These novel findings suggest that loss of Hint1 expression may play a role in the pathogenesis of a subset of human hepatocellular carcinomas. Indeed, in recent unpublished studies we have found that there is an increased frequency of hypermethylation of the promoter region of the Hint1 gene in hepatocellular carcinoma samples from Taiwan when compared to adjacent normal liver tissue or normal liver tissue (unpublished data). Further studies are in progress to determine the clinical relevance of impairments in the expression and function of HINT1 with respect to hepatocellular carcinoma and other types of human cancer.
Acknowledgments
Grant support: Entertainment Industry Foundation-National Colorectal Cancer Research Alliance, the T. J. Martell Foundation, and the National Foundation for Cancer Research (to I.B. Weinstein); National Natural Science Foundation of China 30660184 (to Lin Wang); and National Cancer Institute Grant ES09089 (to Regina M Santella). The costs of publication of this article were defrayed in part by the payment of page charges.
The authors thank Richard Pestell for generously providing the p1745 bp CDI LUC, Kathryn Calame for the E-box LUC, Angie Rizzino for the TGFβ2 CAT and Michele Sawadogo for the pCMV-USF2 plasmids. We also thank Steven Xing for valuable technical support and Barbara Castro for administrative expertise.
Abbreviations used are
- HIT
histidine triad
- HINT1
HIT nucleotide binding protein-1
- FHIT
fragile locus HIT protein
- HA
hemagglutinin
- PKC
protein kinase C
- 5-Azadc
5-azadeoxycytidine
Footnotes
Novelty and Impact of this paper: This paper provides the first evidence that loss of expression of the novel tumor suppressor gene Hint1 due to promoter methylation may play a role in the pathogenesis of a subset of human hepatoma by enhancing the activities of the transcription factors β-catenin, USF2 and NFκB.
This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.Li H, Zhang Y, Su T, Santella RM, Weinstein IB. Hint1 is a haplo-insufficient tumor suppressor in mice. Oncogene. 2006;25:713–21. doi: 10.1038/sj.onc.1209111. [DOI] [PubMed] [Google Scholar]
- 2.Su T, Suzui M, Wang L, Lin CS, Xing WQ, Weinstein IB. Deletion of histidine triad nucleotide-binding protein 1/PKC-interacting protein in mice enhances cell growth and carcinogenesis. Proc Natl Acad Sci U S A. 2003;100:7824–9. doi: 10.1073/pnas.1332160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan BZ, Jefferson AM, Popescu NC, Reynolds SH. Aberrant gene expression in human non small cell lung carcinoma cells exposed to demethylating agent 5-aza-2′-deoxycytidine. Neoplasia. 2004;6:412–9. doi: 10.1593/neo.03490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Zhang Y, Li H, Xu Z, Santella RM, Weinstein IB. Hint1 inhibits growth and activator protein-1 activity in human colon cancer cells. Cancer Res. 2007;67:4700–8. doi: 10.1158/0008-5472.CAN-06-4645. [DOI] [PubMed] [Google Scholar]
- 5.Korsisaari N, Makela TP. Interactions of Cdk7 and Kin28 with Hint/PKCI-1 and Hnt1 histidine triad proteins. J Biol Chem. 2000;275:34837–40. doi: 10.1074/jbc.C000505200. [DOI] [PubMed] [Google Scholar]
- 6.Razin E, Zhang ZC, Nechushtan H, Frenkel S, Lee YN, Arudchandran R, Rivera J. Suppression of microphthalmia transcriptional activity by its association with protein kinase C-interacting protein 1 in mast cells. J Biol Chem. 1999;274:34272–6. doi: 10.1074/jbc.274.48.34272. [DOI] [PubMed] [Google Scholar]
- 7.Lee YN, Razin E. Nonconventional involvement of LysRS in the molecular mechanism of USF2 transcriptional activity in FcepsilonRI-activated mast cells. Mol Cell Biol. 2005;25:8904–12. doi: 10.1128/MCB.25.20.8904-8912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiske J, Huber O. The histidine triad protein Hint1 interacts with Pontin and Reptin and inhibits TCF-beta-catenin-mediated transcription. J Cell Sci. 2005;118:3117–29. doi: 10.1242/jcs.02437. [DOI] [PubMed] [Google Scholar]
- 9.Scholtz B, Kingsley-Kallesen M, Rizzino A. Transcription of the transforming growth factor-beta2 gene is dependent on an E-box located between an essential cAMP response element/activating transcription factor motif and the TATA box of the gene. J Biol Chem. 1996;271:32375–80. doi: 10.1074/jbc.271.50.32375. [DOI] [PubMed] [Google Scholar]
- 10.Kingsley-Kallesen M, Luster TA, Rizzino A. Transcriptional regulation of the transforming growth factor-beta2 gene in glioblastoma cells. In Vitro Cell Dev Biol Anim. 2001;37:684–90. doi: 10.1290/1071-2690(2001)037<0684:TROTTG>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 11.Roth U, Jungermann K, Kietzmann T. Modulation of glucokinase expression by hypoxia-inducible factor 1 and upstream stimulatory factor 2 in primary rat hepatocytes. Biol Chem. 2004;385:239–47. doi: 10.1515/BC.2004.018. [DOI] [PubMed] [Google Scholar]
- 12.Allen RR, Qi L, Higgins PJ. Upstream stimulatory factor regulates E box-dependent PAI-1 transcription in human epidermal keratinocytes. J Cell Physiol. 2005;203:156–65. doi: 10.1002/jcp.20211. [DOI] [PubMed] [Google Scholar]
- 13.Samoylenko A, Roth U, Jungermann K, Kietzmann T. The upstream stimulatory factor-2a inhibits plasminogen activator inhibitor-1 gene expression by binding to a promoter element adjacent to the hypoxia-inducible factor-1 binding site. Blood. 2001;97:2657–66. doi: 10.1182/blood.v97.9.2657. [DOI] [PubMed] [Google Scholar]
- 14.Miller JR, Hocking AM, Brown JD, Moon RT. Mechanism and function of signal transduction by the Wnt/beta-catenin and Wnt/Ca2+ pathways. Oncogene. 1999;18:7860–72. doi: 10.1038/sj.onc.1203245. [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Angelats M, Teeguarden JG, Dragan YP, Pitot HC. Mutational analysis of three tumor suppressor genes in two models of rat hepatocarcinogenesis. Mol Carcinog. 1999;25:157–63. doi: 10.1002/(sici)1098-2744(199907)25:3<157::aid-mc1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 16.Chen TC, Hsieh LL, Ng KF, Jeng LB, Chen MF. Absence of APC gene mutation in the mutation cluster region in hepatocellular carcinoma. Cancer Lett. 1998;134:23–8. doi: 10.1016/s0304-3835(98)00238-9. [DOI] [PubMed] [Google Scholar]
- 17.de La Coste A, Romagnolo B, Billuart P, Renard CA, Buendia MA, Soubrane O, Fabre M, Chelly J, Beldjord C, Kahn A, Perret C. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci U S A. 1998;95:8847–51. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anna CH, Sills RC, Foley JF, Stockton PS, Ton TV, Devereux TR. Beta-catenin mutations and protein accumulation in all hepatoblastomas examined from B6C3F1 mice treated with anthraquinone or oxazepam. Cancer Res. 2000;60:2864–8. [PubMed] [Google Scholar]
- 20.Tsujiuchi T, Tsutsumi M, Sasaki Y, Takahama M, Konishi Y. Different frequencies and patterns of beta-catenin mutations in hepatocellular carcinomas induced by N-nitrosodiethylamine and a choline-deficient L-amino acid-defined diet in rats. Cancer Res. 1999;59:3904–7. [PubMed] [Google Scholar]
- 21.Inagawa S, Itabashi M, Adachi S, Kawamoto T, Hori M, Shimazaki J, Yoshimi F, Fukao K. Expression and prognostic roles of beta-catenin in hepatocellular carcinoma: correlation with tumor progression and postoperative survival. Clin Cancer Res. 2002;8:450–6. [PubMed] [Google Scholar]
- 22.Bosch FX, Ribes J, Cleries R, Diaz M. Epidemiology of hepatocellular carcinoma. Clin Liver Dis. 2005;9:191–211. v. doi: 10.1016/j.cld.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Meier JL, Luo X, Sawadogo M, Straus SE. The cellular transcription factor USF cooperates with varicella-zoster virus immediate-early protein 62 to symmetrically activate a bidirectional viral promoter. Mol Cell Biol. 1994;14:6896–906. doi: 10.1128/mcb.14.10.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzui M, Masuda M, Lim JT, Albanese C, Pestell RG, Weinstein IB. Growth inhibition of human hepatoma cells by acyclic retinoid is associated with induction of p21(CIP1) and inhibition of expression of cyclin D1. Cancer Res. 2002;62:3997–4006. [PubMed] [Google Scholar]
- 25.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–10. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 26.Lu T, Burdelya LG, Swiatkowski SM, Boiko AD, Howe PH, Stark GR, Gudkov AV. Secreted transforming growth factor beta2 activates NF-kappaB, blocks apoptosis, and is essential for the survival of some tumor cells. Proc Natl Acad Sci U S A. 2004;101:7112–7. doi: 10.1073/pnas.0402048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong CM, Fan ST, Ng IO. beta-Catenin mutation and overexpression in hepatocellular carcinoma: clinicopathologic and prognostic significance. Cancer. 2001;92:136–45. doi: 10.1002/1097-0142(20010701)92:1<136::aid-cncr1301>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 28.Weiske J, Albring KF, Huber O. The tumor suppressor Fhit acts as a repressor of beta-catenin transcriptional activity. Proc Natl Acad Sci U S A. 2007;104:20344–9. doi: 10.1073/pnas.0703664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Payton SG, Liu M, Ge Y, Matherly LH. Transcriptional regulation of the human reduced folate carrier A1/A2 promoter: Identification of critical roles for the USF and GATA families of transcription factors. Biochim Biophys Acta. 2005;1731:115–24. doi: 10.1016/j.bbaexp.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Ge Y, Jensen TL, Matherly LH, Taub JW. Physical and functional interactions between USF and Sp1 proteins regulate human deoxycytidine kinase promoter activity. J Biol Chem. 2003;278:49901–10. doi: 10.1074/jbc.M305085200. [DOI] [PubMed] [Google Scholar]
- 31.Cogswell JP, Godlevski MM, Bonham M, Bisi J, Babiss L. Upstream stimulatory factor regulates expression of the cell cycle-dependent cyclin B1 gene promoter. Mol Cell Biol. 1995;15:2782–90. doi: 10.1128/mcb.15.5.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.North S, Espanel X, Bantignies F, Viollet B, Vallet V, Jalinot P, Brun G, Gillet G. Regulation of cdc2 gene expression by the upstream stimulatory factors (USFs) Oncogene. 1999;18:1945–55. doi: 10.1038/sj.onc.1202506. [DOI] [PubMed] [Google Scholar]
- 33.Pawar SA, Szentirmay MN, Hermeking H, Sawadogo M. Evidence for a cancer-specific switch at the CDK4 promoter with loss of control by both USF and c-Myc. Oncogene. 2004;23:6125–35. doi: 10.1038/sj.onc.1207806. [DOI] [PubMed] [Google Scholar]
- 34.Sirito M, Lin Q, Deng JM, Behringer RR, Sawadogo M. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc Natl Acad Sci U S A. 1998;95:3758–63. doi: 10.1073/pnas.95.7.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buck MB, Knabbe C. TGF-beta signaling in breast cancer. Ann N Y Acad Sci. 2006;1089:119–26. doi: 10.1196/annals.1386.024. [DOI] [PubMed] [Google Scholar]
- 36.Govinden R, Bhoola KD. Genealogy, expression, and cellular function of transforming growth factor-beta. Pharmacol Ther. 2003;98:257–65. doi: 10.1016/s0163-7258(03)00035-4. [DOI] [PubMed] [Google Scholar]
- 37.Vagenas K, Spyropoulos C, Gavala V, Tsamandas AC. TGFbeta1, TGFbeta2, and TGFbeta3 protein expression in gastric carcinomas: correlation with prognostics factors and patient survival. J Surg Res. 2007;139:182–8. doi: 10.1016/j.jss.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Grau AM, Datta PK, Zi J, Halder SK, Beauchamp RD. Role of Smad proteins in the regulation of NF-kappaB by TGF-beta in colon cancer cells. Cell Signal. 2006;18:1041–50. doi: 10.1016/j.cellsig.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 39.Wei YY, Chen YJ, Hsiao YC, Huang YC, Lai TH, Tang CH. Osteoblasts-derived TGF-beta1 enhance motility and integrin upregulation through Akt, ERK, and NF-kappaB-dependent pathway in human breast cancer cells. Mol Carcinog. 2008;47:526–37. doi: 10.1002/mc.20411. [DOI] [PubMed] [Google Scholar]
- 40.Descargues P, Sil AK, Sano Y, Korchynskyi O, Han G, Owens P, Wang XJ, Karin M. IKKalpha is a critical coregulator of a Smad4-independent TGFbeta-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc Natl Acad Sci U S A. 2008;105:2487–92. doi: 10.1073/pnas.0712044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu T, Sathe SS, Swiatkowski SM, Hampole CV, Stark GR. Secretion of cytokines and growth factors as a general cause of constitutive NFkappaB activation in cancer. Oncogene. 2004;23:2138–45. doi: 10.1038/sj.onc.1207332. [DOI] [PubMed] [Google Scholar]
- 42.Boon RA, Fledderus JO, Volger OL, van Wanrooij EJ, Pardali E, Weesie F, Kuiper J, Pannekoek H, ten Dijke P, Horrevoets AJ. KLF2 suppresses TGF-beta signaling in endothelium through induction of Smad7 and inhibition of AP-1. Arterioscler Thromb Vasc Biol. 2007;27:532–9. doi: 10.1161/01.ATV.0000256466.65450.ce. [DOI] [PubMed] [Google Scholar]
- 43.Viollet B, Lefrancois-Martinez AM, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J Biol Chem. 1996;271:1405–15. doi: 10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]
- 44.Yan S, Sloane BF. Isolation of a novel USF2 isoform: repressor of cathepsin B expression. Gene. 2004;337:199–206. doi: 10.1016/j.gene.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Vallet VS, Casado M, Henrion AA, Bucchini D, Raymondjean M, Kahn A, Vaulont S. Differential roles of upstream stimulatory factors 1 and 2 in the transcriptional response of liver genes to glucose. J Biol Chem. 1998;273:20175–9. doi: 10.1074/jbc.273.32.20175. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh AK, Datta PK, Jacob ST. The dual role of helix-loop--helix-zipper protein USF in ribosomal RNA gene transcription in vivo. Oncogene. 1997;14:589–94. doi: 10.1038/sj.onc.1200866. [DOI] [PubMed] [Google Scholar]
- 47.Cable PL, Wilson CA, Calzone FJ, Rauscher FJ, 3rd, Scully R, Livingston DM, Li L, Blackwell CB, Futreal PA, Afshari CA. Novel consensus DNA-binding sequence for BRCA1 protein complexes. Mol Carcinog. 2003;38:85–96. doi: 10.1002/mc.10148. [DOI] [PubMed] [Google Scholar]