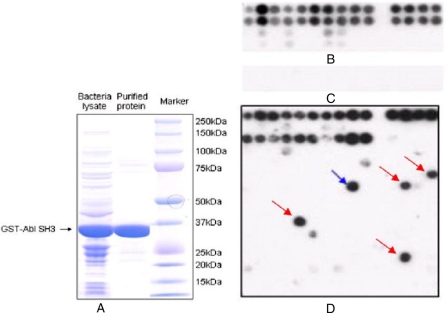
Fig. 2.
Peptide array experiments to validate the computational predictions. A, expression and purification of GST-Abl SH3 fusion protein. The fusion protein was expressed in E. coli BL21 (DE3) and purified on glutathione-agarose. B, control array probed with GST-Abl SH3 followed by anti-GST antibody. The array contained 30 control peptides in duplicates: 15 known binders of the Abl SH3 domain (the first two rows), 10 binders for other domains but not the Abl SH3 domain (the first 10 peptides in the next two rows), and five random peptides that are presumably non-binders. C, control array probed with anti-GST antibody only. The array contained the 30 control peptides mentioned above. D, production array probed with GST-Abl SH3 followed by anti-GST antibody. The array contained the 30 control peptides mentioned above (first two rows) and 210 testing peptides: 10 known binders (the first 10 peptides in the third row) and 200 random peptides with the PXXP motif selected from the human proteome. Five binders (blue and red arrows) were identified from the 200 random peptides, one of which (blue arrow) was also identified as a true binder by the MIEC-SVM method.