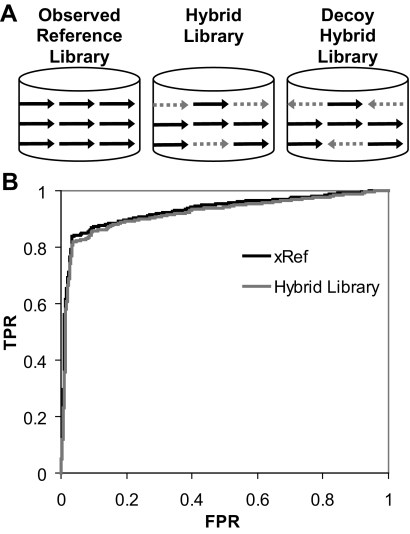
Fig. 1.
Performance of simulated MS/MS spectra within a reference library. A, strategy for generating spectral libraries to test simulated spectra. Spectra in the reference library (xRef) that corresponded to peptides in 49 standard proteins were replaced by simulated spectra of the same peptide ions generated by MassAnalyzer, producing the hybrid library. A decoy hybrid library was created by replacing xRef spectra with simulated spectra generated against peptide sequences reversed to read from C to N terminus, holding the C-terminal amino acid constant to maintain the tryptic nature of the sequence. Only the simulated spectra correspond to inverted sequences and thus represent decoy targets. B, ROC plots of results using xRef or the hybrid library to search an LC-MS/MS data set collected for a tryptic digest of the 49 standard proteins (ABRF data set). Each plotted point represents the true positive rate (TPR) as the proportion of the MS/MS assigned correctly (as ABRF proteins), when scoring above the threshold at each plotted point, divided by the total number of MS/MS assigned correctly in this data set. The false positive rate (FPR; (FP/(TN + FP))) is the proportion of incorrect assignments (not assigned to any ABRF protein) above the threshold at each plotted point divided by the total number of incorrect assignments. The algorithm for calculating these values is given in the supplemental data.