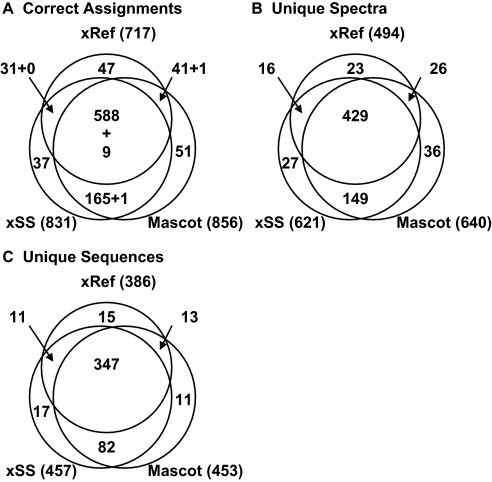
Fig. 3.
Search results for the ABRF data set using X!Hunter or Mascot. Searches with X!Hunter used a reference library of observed spectra (xRef) or the full simulated spectral library (xSS), whereas searches with Mascot used the IPI protein database (“Mascot”). A, summary of all identified MS/MS. B, summary of the identified unique spectra, which count peptide ions with distinct sequence and/or charge state. C, summary of the identified unique peptide sequences. The total numbers of assignments to ABRF proteins for each search strategy are given in parentheses. In a few cases, more than one ABRF assignment was made to the same spectrum, and these are indicated as a second number (e.g. “+ 9” in A, region showing overlap between all three searches). Manual validation confirmed that both assignments were correct because the MS/MS was a chimera of two different parent ions captured in one MS/MS isolation event. To compare xRef and xSS, MS/MS derived from non-tryptic sequences were removed from the ABRF data set. Only peptides with m/z >900 Da and that were derived from singly, doubly, or triply charged parent ions were included.