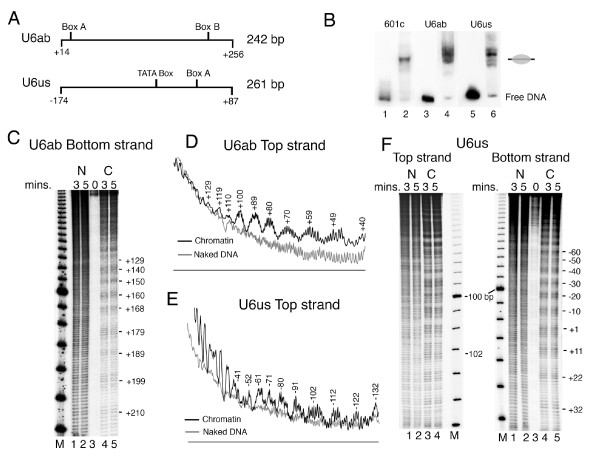
Figure 3.
Nucleosomes on the SNR6 gene are rotationally positioned. (A) Schematic representation of the selected gene regions, PCR-amplified from p-539H6 with their names and sizes is given. (B) Gel mobility shift assay of chromatin assembled over end-labeled 601c, SNR6ab and SNR6us DNA fragments (lanes 2, 4 and 6, respectively). Lanes 1, 3 and 5 show free DNA controls. Gray oval on the left-hand side marks the position of the centrally positioned nucleosome. (C) to (F) Hydroxyl radical cleavage of the DNA. Numbers identify the cleavage peaks of chromatin in base pairs. (C) and (F) Digestion times are given in minutes; 0 min. represents undigested DNA. Cleavage pattern of the chromatin samples, C, are shown in the lanes 3 and 4 for the top strand gel and lanes 4 and 5 for bottom strand gels. Lanes 1 and 2 show digestion pattern of naked DNA (N), while a 10 bp DNA ladder (Invitrogen) was end-labeled and used as size marker in the lanes M. (D) Profile of the digestion pattern of the top strand of SNR6ab chromatin. (E) Profile of the digestion pattern of the top strand of SNR6us chromatin from the gel shown in panel (F).