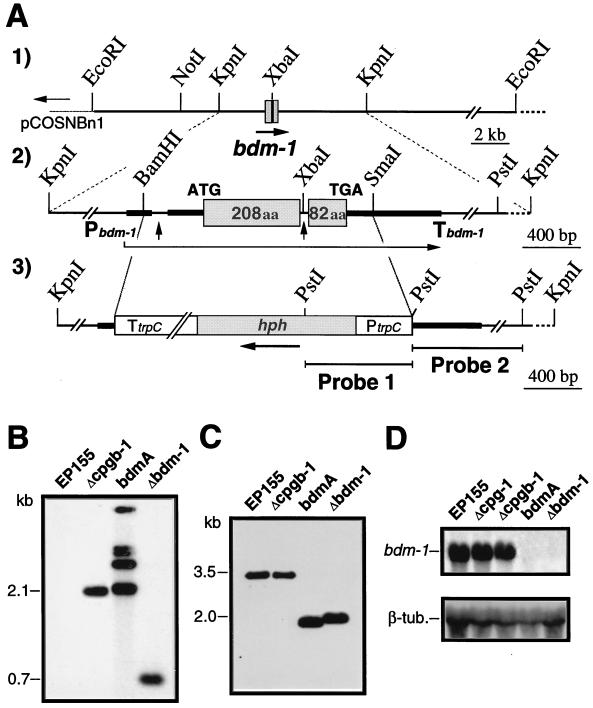
Figure 3.
Complementation mapping and disruption of bdm-1. (A1) Restriction map of cosmid clone rescued from the chromosome of a bdmA complemented transformant. (A2) Detailed organization of bdm-1 in the context of the 6.5-kb KpnI bdmA complementing fragment. The bdm-1 promoter and terminator are indicated by Pbdm-1 and Tbdm-1, respectively. The protein-coding regions are represented by boxes. Thick solid lines denote 5′ and 3′ noncoding region. Two introns are marked by vertical arrows. (A3) DNA fragment used to disrupt bdm-1. The BamHI–SmaI fragment within the bdm-1 structural gene was replaced with a hygromycin-resistance gene (hph) cassette that contained the A. nidulans TrpC promoter and terminator, PtrpC and TtrpC, respectively (49). A 7.3-kb fragment released by digestion with KpnI was used directly for transformation of C. parasitica strain EP155 spheroplasts. (B) Southern blot analysis of PstI-digested genomic DNA (10 μg) isolated from wild-type C. parasitica strain EP155, Gβ disruptant Δcpgb-1, bdmA, and bdm-1 disruptant Δbdm-1 by using probe 1 (specific for the hph gene) shown in A (3) above, under conditions described in ref. 18. Sizes of expected hybridization bands are indicated at the left. (C) Southern blot analysis of the same strains listed in B by using probe 2 shown in A (3), which is specific for bdm-1. (D) Northern blot analysis of RNA (15 μg) isolated from the same strains shown in C and the cpg-1 disruptant Δcpg-1 by using the entire coding region of bdm-1 cDNA as probe as described in ref. 18.