Figure 3.
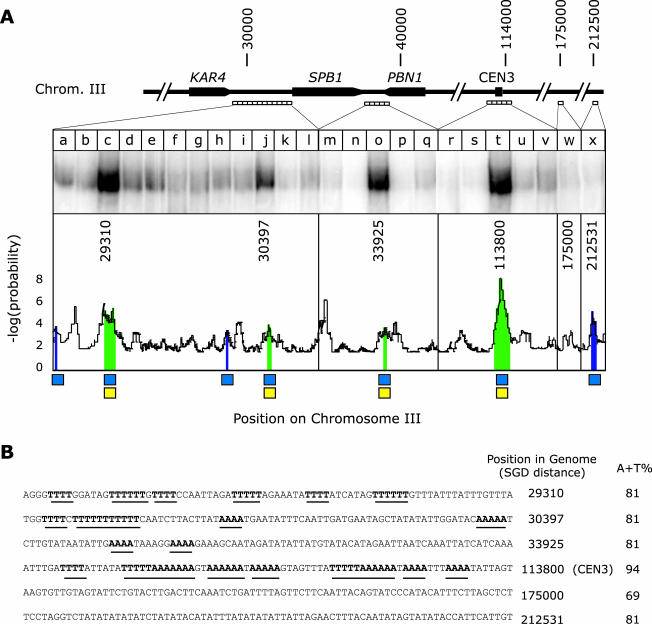
Binding of recombinant Ndc10p to sequences on Chromosome III arms and in CEN3. (A) DNA probes in 200-base pair increments were generated across the indicated regions in the KAR4-PBN1 interval and analyzed for binding to purified recombinant Ndc10p. Distances are indicated in nucleotides from the left telomere of Chromosome III. The graph shows the product of the log probability of the A-T composition in successive 80-base pair intervals and the log probability of the number of stretches containing four or more adenines or thymines in a row. To find potential Ndc10p binding sites, sequences were sought in which the A-T composition exceeded 80% (blue boxes) and the number of poly-A/poly-T stretches was significantly above average for the genome (yellow boxes). Ndc10p binding was predicted to occur on sequences in which both of these thresholds had been exceeded (green regions). (B) Sequences of six 80-base pair windows in Chromosome III analyzed for Ndc10p binding in vitro. Position refers to the number of the first nucleotide in the window relative to the left telomere of Chromosome III (Cherry et al., 1997) and “A + T” % to the percentage of adenine plus thymine. Stretches of A and T are emphasized in bold and underlined. Sequence 113800 is CDEII of CEN3; 175000 (lane w) represents a randomly chosen genomic location; 212531 is a site of high A-T content but lacking poly-A/poly-T stretches.