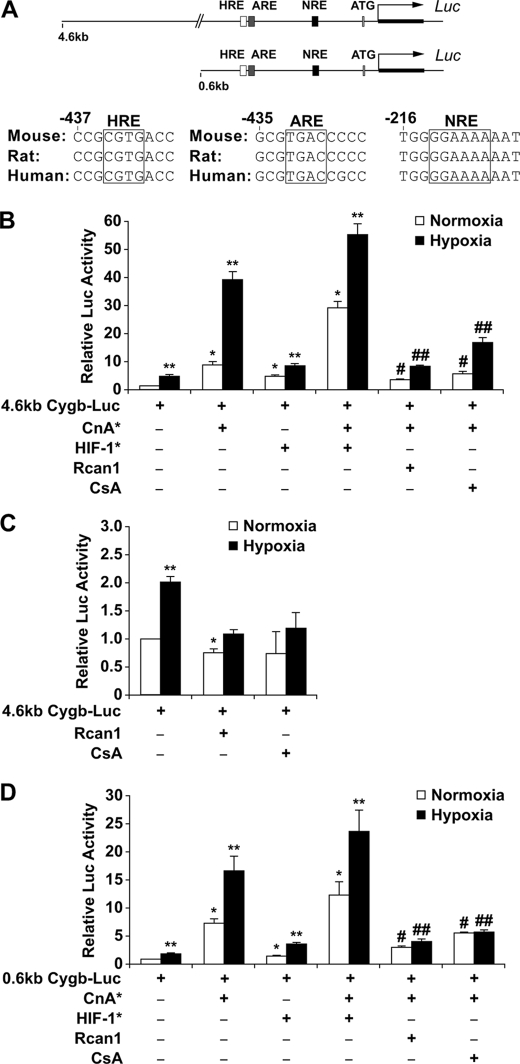
FIGURE 2.
Calcineurin regulates the transcriptional activity of the 5′ upstream region of the cytoglobin gene. A, depiction of two regions (4.6- and 0.6-kb fragments) upstream of the 5′-end of the Cygb gene illustrates the evolutionarily conserved motifs for HIF-1, AP-1, and NFAT located within the first 600 base pairs. B, the transcriptional activity of the 4.6-kb Cygb-Luc plasmid was assessed in C2C12 myoblasts. Co-transfection with a CnA* expression plasmid resulted in a 39.3 ± 3.0-fold induction of transcriptional activity under hypoxic conditions. This response was significantly blunted by inhibitors of CnA* (Rcan1 or CsA). Co-transfection with a HIF* expression plasmid increased both basal and CnA*-stimulated transcription. C, inhibition of endogenous calcineurin activity (by overexpressing Rcan1 or exposure to CsA) in C2C12 cells resulted in complete attenuation of transcriptional activation of the 4.6-kb Cygb-Luc reporter under both normoxic and hypoxic conditions. D, transcriptional assays of the 0.6-kb Cygb-Luc plasmid transfected into C2C12 cells revealed a pattern of activation similar to the 4.6-kb Cygb-Luc reporter. Co-transfection with a CnA* expression plasmid resulted in a 16.8 ± 2.6-fold induction of transcriptional activity under hypoxic conditions. This response was also minimally augmented with HIF-1* but was significantly blunted by inhibitors of calcineurin (Rcan1 and CsA). *, p < 0.005 normoxic experimental cells versus normoxic control cells transfected with only the promoter reporter plasmid; **, p < 0.005 hypoxic experimental cells versus normoxic control cells transfected with only the promoter reporter plasmid; #, p < 0.01 normoxic experimental cells with inhibited calcineurin activity versus normoxic cells overexpressing CnA*; ##, p < 0.01 hypoxic experimental cells with inhibited calcineurin activity versus hypoxic cells overexpressing CnA* (n = 9 in each group).