Figure 3.
Mutation in SLC33A1 Causes HSP
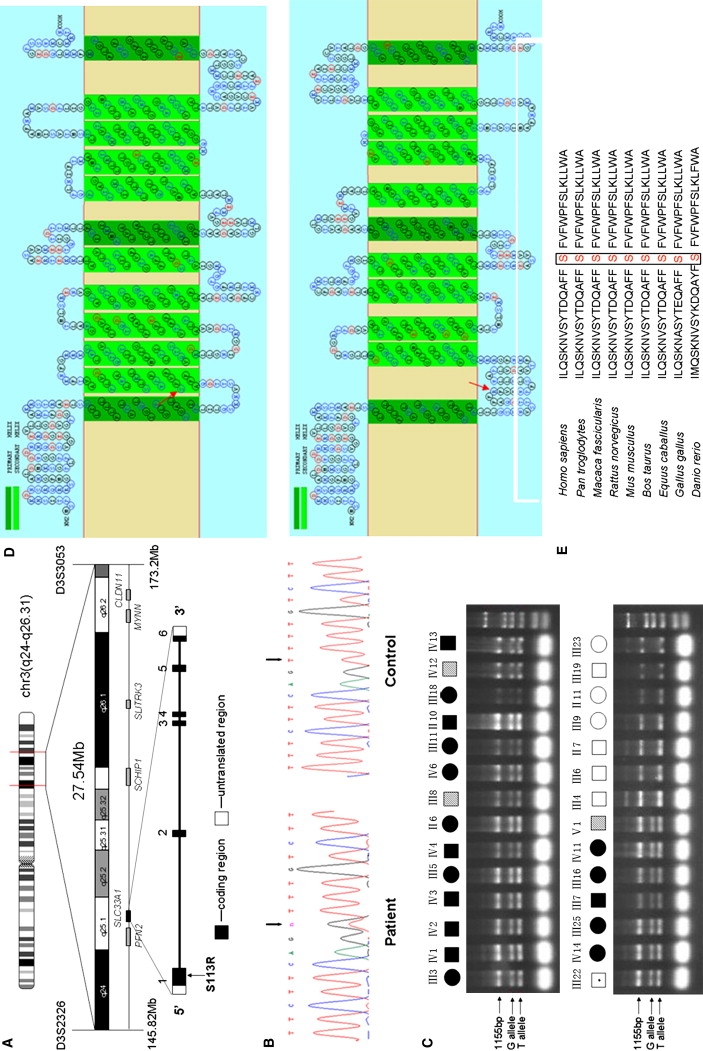
(A) The candidate region of 27.54 Mb between D3S2326 and D3S3053. The candidate genes screened are shown at the bottom. The location of SLC33A1 is highlighted.
(B) Partial sequence chromatograms of exon 1 of SLC33A1 in a patient and a healthy control. The arrows mark the position of the SLC33A1 mutation. Sequence analysis was done on an ABI 3100 automated sequencer (Applied Biosystems).
(C) Confirmation of the c.339T→G mutation by allele-specific tetra-primer PCR assay. All definitely affected and mildly affected individuals exhibited the G allele. Primers sequences are as follows: forward outer primer 5′-GTGCCCTTATCGCTCTGA-3′, reverse outer primer 5′-GTGTTATTTGATGGGTTGC-3′, forward inner primer (G allele) 5′-TTAGCTATACAGACCAAGCTTTCTTCGGG-3′, and reverse inner primer (T allele) 5′-TTGAGACTGAAGGGCCAAAAGACAGAA-3′.
(D) Topological prediction of the mutant SLC33A1 protein, with use of the SOSUI program. The S113R mutation resulted in the loss of the original second TM motif and reversed the orientation of all domains, starting from 113. Upper panel, normal SLC33A1; bottom panel, mutant SLC33A1 (S113R). The mutated amino acid is indicated by the red arrow.
(E) Amino acid sequence alignment of human SLC33A1 and orthologs from other species, showing phylogenetic conservation of S113. The sequences were retrieved from the Entrez protein database and aligned to each other with the use of Clustal W.
