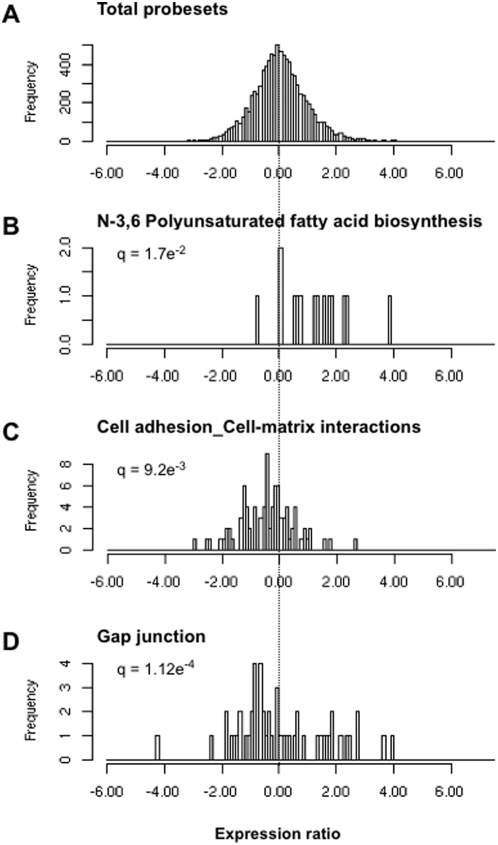
Figure 3. Gene set enrichment results.
Distribution of 2D normalized z-scores (NZ2D) for (A) all cDNA clones used in this screen and clones assigned to the (B) N-3,6 Polyunsaturated fatty acid biosynthesis, (C) Cell Adhesion / Cell Matrix Interaction and (D) Gap Junction pathways. The rightward shift of NZ2D scores among fatty acid synthesis genes (B) relative to background (A) indicates that overexpression of genes in this pathway on balance tend to activate SREBP transcriptional activity, whereas the leftward shift in (C) indicates that the cell adhesion/cell matrix tend to inhibit SREBP transcriptional activity in this screen. Gap junction genes were spread on either side of the median (D) indicating that a sub-set of gap junction genes activated the SREBP pathway, while others repressed it.