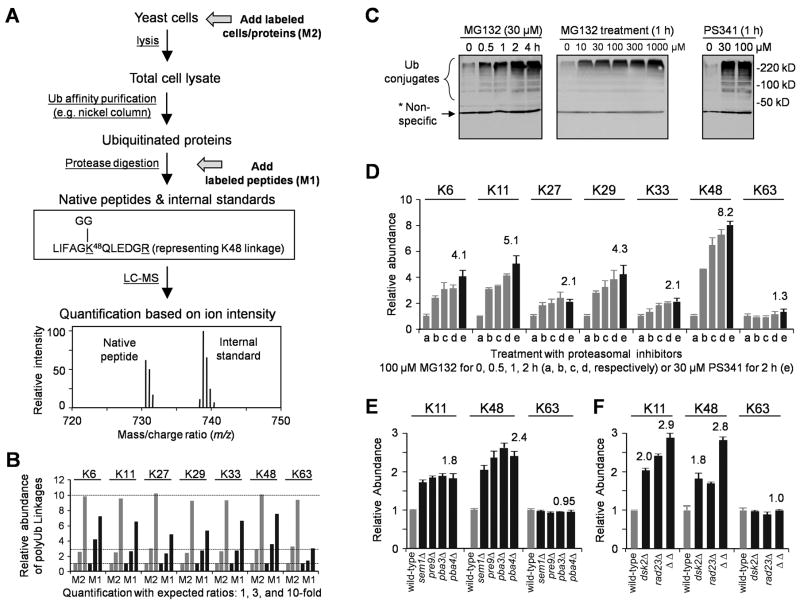
Figure 1.
Inhibition of the Ub-proteasome system increases the level of all non-K63 linked polyUb chains.
(A) Schematic of quantitative MS. In Method 1 (M1), isotope-labeled labeled peptides are used as standards that are added during trypsin digestion. In Method 2 (M2), labeled cells/proteins are spiked in after cell harvest, but before cell lysis, minimizing variations in sample processing. In addition, labeled proteins can be added in any steps between cell lysis and trypsin digestion as internal standards.
(B) Using labeled cells/proteins (M2, grey) instead of peptides (M1, black) reduced quantitative variations. Three yeast lysates with different amounts of polyUb chains (1x, 3x and 10x) were processed in parallel, and the data were normalized to the result of the 1× sample.
(C) Proteasome inhibitor treatment caused accumulation of Ub conjugates in a dose- and time-dependent manner. Strain JMP001 (pdr5Δ) expressing His-myc-Ub was treated and harvested for immunoblotting with myc antibodies.
(D) Distinct polyUb chain linkages were measured by MS, shown as mean ± SEM.
(E–F) Yeast strains with mutations in Ub-proteasome system raised K11 and K48 linkages but not K63-linked chains. Data are represented as mean and SEM.